Summary
Human lymphoid malignancies inherit gene expression networks from their normal B-cell counterpart and co-opt them for their own oncogenic purpose, which is usually governed by transcriptional factors and signaling pathways. These transcriptional factors and signaling pathways are precisely regulated at multiple steps, including ubiquitin modification. With a function involved in almost all cellular events, protein ubiquitination plays a role in many human diseases. In the past few years, multiple studies have expanded the role of ubiquitination in the genesis of diverse lymphoid malignancies. Here we discuss our current understanding of both proteolytic and nonproteolytic functions of the protein ubiquitination system and describe how it is involved in the pathogenesis of human lymphoid cancers. Lymphoid-restricted ubiquitination mechanisms, including ubiquitin E3 ligases and deubiquitinating enzymes, provide great opportunities for the development of targeted therapies for lymphoid cancers.
Keywords: ubiquitination, E3 ligase, lymphoma, multiple myeloma, ABC DLBCL
Introduction
It is becoming clear that the different types of human lymphoid cancer derive much of their biology from the various stages of normal B-cell differentiation from which they originate (1). While pre-germinal center mature B cells appear to be the origin of mantle cell lymphoma (MCL) and chronic lymphocytic leukemia (CLL), the majority of non-Hodgkin’s lymphomas as well as multiple myeloma originate from germinal center B cells or from B cells that have passed through this developmental stage. To understand the oncogenic mechanisms of these malignancies, gene expression profiling has proven extremely instructive. Based on gene expression signatures, three types of non-Hodgkin’s lymphoma, follicular lymphoma (FL), Burkitt lymphoma (BL), and the germinal center B cell–like (GCB) subtype of diffuse large B cell lymphoma (DLBCL), appear to arise from germinal center B cells, whereas the activated B cell-like (ABC) subtype of DLBCL appears to arise from a post–germinal center B cell (2–4). Primary mediastinal B-cell lymphoma (PMBL), the third DLBCL subtype, may originate from a rare thymic B-cell subpopulation, as does Hodgkin’s lymphoma, which has a highly similar gene expression profile (5, 6). Multiple myeloma, although phenotypically resembling plasma cells, likely also originates within the germinal center since several recurrent chromosomal translocations are caused by DNA breaks created by the enzyme activation-induced cytidine deaminase (AID), which is preferentially expressed in the germinal center (7).
Given these varied origins, it is perhaps not surprising that each type of B-cell malignancy acquires characteristic genomic abnormalities – mutations, amplifications, deletions and translocations – that promote its malignant behavior (reviewed in 1). Many of these aberrations constitutively activate signaling pathways that are used in an inducible fashion by normal B cells. The result of this constitutive signaling is the alteration of transcriptional networks that promote proliferation and survival, which are governed by master regulatory transcription factors. In some cases these master regulators are directly dysregulated by chromosomal rearrangements that target their loci, as occurs with Bcl6, Irf4, and c-Myc.
A key concept in cancer biology is ‘oncogene addiction’ in which cancer cells are strikingly dependent on a genetically altered oncoprotein, making them ideal drug targets (8). However, cancer cells can be just as addicted to unmutated proteins as an indirect consequence of their altered regulatory networks, a phenomenon that has been generically dubbed ‘non-oncogene addition’ (9). Broadly, non-oncogene addiction can be subdivided into three categories. First, cancer cells can rely on key master transcription factors that are also utilized by the normal cell lineage from which they derive, which have been termed ‘lineage survival oncogenes’ (10). A second type of non-oncogene addition is established when a mutant signaling protein engages an entire pathway, causing the cancer cells to become critically dependent on wildtype proteins in the pathway. Finally, the abnormal cellular physiology of cancer can invoke various ‘stress’ responses that then become essential to the survival of the cancer cells. The present review focuses on non-oncogene addiction of lymphoid cancers and the role of the ubiquitin system in this process.
A prime example of non-oncogene addiction is provided by the family of transcription factors known collectively as nuclear factor κB (NF-κB). Perhaps the most common approach taken by lymphoid malignancies to avoid cell death is the constitutive activation of the NF-κB pathway (reviewed in 11). While NF-κB family members themselves are usually not targeted by genetic alterations, various signal transduction pathways that become constitutively actively as a consequence of oncogenic events lead to the nuclear translocation of NF-κB transcription factors and subsequent activation of a transcriptional program that promotes cellular survival and proliferation. These oncogenic signaling pathways are regulated at many levels by the protein ubiquitination system.
Oncogenic signaling in ABC DLBCL
NF-κB is activated by constitutive activity of various upstream signaling pathways in the activated B-cell like (ABC) subtype of DLBCL. This subtype was so-named because its gene expression profile resembles that of normal B cells that have been activated by antigen engagement of the B-cell receptor (BCR) (2). The BCR triggers several downstream signaling responses that promote proliferation and survival of normal B cells during development and immune responses, but these responses must be tightly regulated to maintain homeostasis. In particular, the BCR initiates a cascade of signaling proteins that work in concert to activate IκB kinase β (IKKβ), the central regulatory kinase of the ‘classical’ NF-κB pathway (reviewed in 12). Lymphocytes utilize a number of other receptor systems to activate NF-κB, including various TNF receptor family members and the Toll-like receptors. Virtually all ABC DLBCL tumors have a gene expression signature indicative of NF-κB pathway activation, and cell line models of ABC DLBCL have active IKKβ, implying that one or more signaling pathways constitutively engage NF-κB in this lymphoma subtype (13).
The oncogenic pathways leading to constitutive activation of NF-κB signaling ABC DLBCL were uncovered using a combination of RNA interference (RNAi) screening and high-throughput resequencing of the genome and transcriptome of ABC DLBCL tumors (reviewed in 1). The first insights came from RNAi screens that showed that the survival of several ABC DLBCL cell lines depends on a trio of signaling adapters: CARD11, MALT1, and BCL10 (14). These proteins form the so-called ‘CBM’ complex that is involved in the antigen-dependent activation of NF-κB (reviewed in 15). Recent analysis of the structural architecture of the CBM signalosome revealed a filamentous morphology, supporting a signaling model in which CARD11 nucleates long homopolymers of BCL10 that recruit MALT1 and IKK, leading to NF-κB activation (16). During normal BCR signaling, CARD11 is activated by phosphorylation of its linker domain by protein kinase C β (PKCβ), which relieves the inhibitory influence of the linker domain on the CARD11 coiled-coil domain. The CARD11 coiled-coil domain and the CARD domain mediate CARD11 multimerization, thereby increasing BCL10 polymerization. The BCL10 filaments most likely serve as platforms to recruit other components of the signalosome, including the protease MALT1, E3 ubiquitin ligases (discussed later), and IKK, leading to IKK activation. In ~10% of ABC DLBCL cases, CARD11 acquires activating mutations in the coiled-coil domain that spontaneously relieve inhibition by the linker domain without the need for PKCβ phosphorylation, causing the formation of large CARD11 aggregates in the cytoplasm that recruit and activate IKK (17).
In the majority of ABC DLBCL cases, CARD11 is not mutated, yet the tumor cells still depend on CARD11 for NF-κB activation and survival, indicating the involvement of upstream BCR signaling (18). Indeed, depletion of BCR subunits (IgM, Igκ/λ, CD79A, CD79B) or signal components upstream of CARD11 in the BCR pathway (BTK, PLCγ2, PKCβ) is toxic to ABC DLBCL lines but not to GCB DLBCL lines (18). The constitutive activity of the BCR in ABC DLBCL was christened ‘chronic active BCR signaling’ to emphasize its similarity to engagement of the BCR by antigen in normal B cells (18). Over 20% of ABC DLBCL tumors harbor somatic mutations targeting the BCR subunits CD79A and CD79B, whereas other lymphoma subtypes have few if any BCR mutations (18). These CD79A and CD79B mutations alter the immunoreceptor tyrosine-based activation motifs (ITAMs), which are required for proximal events in BCR signaling. The mutant CD79A and CD79B isoforms potentiate BCR signaling by increasing cell surface expression of the BCR and by limiting negative feedback by Lyn kinase (18).
In addition to chronic active BCR signaling, a signaling cascade mediated by the adapter molecule MyD88 is essential for NF-κB activity and cell survival in a large subset of ABC DLBCL cases (19). ABC DLBCL tumors harbor MYD88 mutations in 39% of cases, making MYD88 the most frequently mutated oncogene in this lymphoma subtype. Almost all MYD88 mutations affect the TIR domain, which mediates interactions between MyD88 and the Toll-like receptors in innate immune responses. The most predominant MYD88 mutation, L265P, occurs in 29% of cases and spontaneously coordinates a signaling complex consisting of the kinases IRAK1 and IRAK4 (19). This particular MyD88 mutant has been identified as a recurrent oncogene in several other lymphoid malignancies, including marginal zone lymphomas, chronic lymphocytic leukemia, and Waldenstrom’s macroglobulinemia (19–21). Depletion of MyD88, IRAK1, or IRAK4 is lethal for ABC DLBCL lines (19). MyD88 mutants promote ABC DLBCL viability by strongly activating NF-κB and by stimulating autocrine IL-6 and IL-10 signaling through JAK kinases and STAT3. In addition, MyD88 signaling also promotes the production of type I interferon in ABC DLBCL, which is less adaptive and must be kept under strict regulatory control (see below).
Protein ubiquitination system
Protein ubiquitination is an essential and pervasive mechanism by which eukaryotic cells regulate responses to multiple cellular stimuli and stresses (22). The function and turnover of a multitude of cellular proteins is altered by the ATP-dependent covalent attachment of ubiquitin, a highly conserved, 76-amino-acid peptide. Ubiquitin is attached by an isopeptide bond linkage to epsilon amino groups of lysines in the substrate protein. Occasionally, ubiquitin can also be conjugated to the amino-terminal methionine residues of substrate proteins.
Protein ubiquitination proceeds by a three-step cascade mechanism (23), which starts with ATP-dependent ubiquitin activation by a ubiquitin-activating enzyme (E1), followed by the transfer of an activated ubiquitin to a cysteine residue within an ubiquitin-conjugating enzyme (E2, also known as UBC), and ending with the conjugation of ubiquitin to a target protein through the activity of a ubiquitin-protein ligase (E3). This cascade can proceed repeatedly, leading to the formation of polyubiquitin chains. There are seven internal lysines in ubiquitin (K6, K11, K27, K29, K33, K48, and K63), each of which can be used to form an isopeptide linkage with the carboxy-terminal glycine of another ubiquitin peptide, generating polyubiquitin chains with distinct structures. More recently, a novel form of polyubiquitin chain has been discovered in which linkages are formed between the amino-terminal methionine of one ubiquitin moiety and the carboxy-terminal glycine of another, resulting in a head-to-tail polymer that is referred to as linear, or M1-linked, polyubiquitin (24).
The human genome encodes two ubiquitin E1 enzymes, 38 E2 enzymes, and more than 600 E3 ubiquitin ligases. An E3 ubiquitin ligase, sometimes together with an E2, determines substrate specificity. E3 ubiquitin ligases constitute a large protein family that can be divided into four categories based on their E3 ligase domains: HECT (homology to E6AP C-terminus) domain, RING (really interesting new gene) domain, U Box domain, and RBR (ring between ring fingers) domain. The HECT domain E3 ubiquitin ligases contain an active-site cysteine that can accept ubiquitin from an E2 enzyme and transfer the ubiquitin to a target protein. In contrast, the RING and U Box domain E3 ubiquitin ligases do not contain a conventional enzyme active site but instead promote ubiquitination by binding to both protein substrates and E2 enzymes, facilitating the transfer of ubiquitin to the substrate. The RBR family of E3 ligases combine features of both the HECT and RING domain proteins in that they have an active site cysteine that can be charged with ubiquitin and also recruit E2 enzymes carrying ubiquitin. Ubiquitination can be reversed by the action deubiquitinating enzymes (DUBs) (also known as isopeptidases) (25).
Ubiquitination was first described as a signal for proteasome-dependent degradation of the modified protein. Now it is clear that different forms of ubiquitination and the topology of polyubiquitin chains have distinct cellular functions. The conjugation of a single ubiquitin moiety to one or more lysine residues in protein substrate, termed monoubiquitination and multiubiquitination respectively, does not typically lead to proteasomal degradation. The functions of protein monoubiquitination are diverse but include chromatin remodeling, vesicle trafficking and protein subcellular localization. The attachment of single ubiquitin moieties can either promote or inhibit the interaction of the modified protein with other proteins, thus changing the cellular response to certain stresses.
As discussed, polyubiquitin chains can utilize any one of seven internal lysines of ubiquitin, thus generating structural diversity that allows proteins with ubiquitin-binding domains to discriminate between these different linkages (26). K48-linked polyubiquitination, with a chain of at least 4 ubiquitins, targets proteins for proteasome-dependent degradation (27). Similarly, K11-linked polyubiquitination can act as a signal for proteasomal degradation (28). By contrast, K63-linked polyubiquitin chains do not promote proteolysis, and are instead involved in kinase activation, DNA damage responses, endocytosis and signal transduction (29, 30). Linear, or M1-linked, polyubiquitination appears to have an exclusive function in regulating various signaling pathways leading to NF-κB (24, 31–33). Thus far, the roles of K6-, K27-, K29-, and K33-linked polyubiquitin chains have not been delineated.
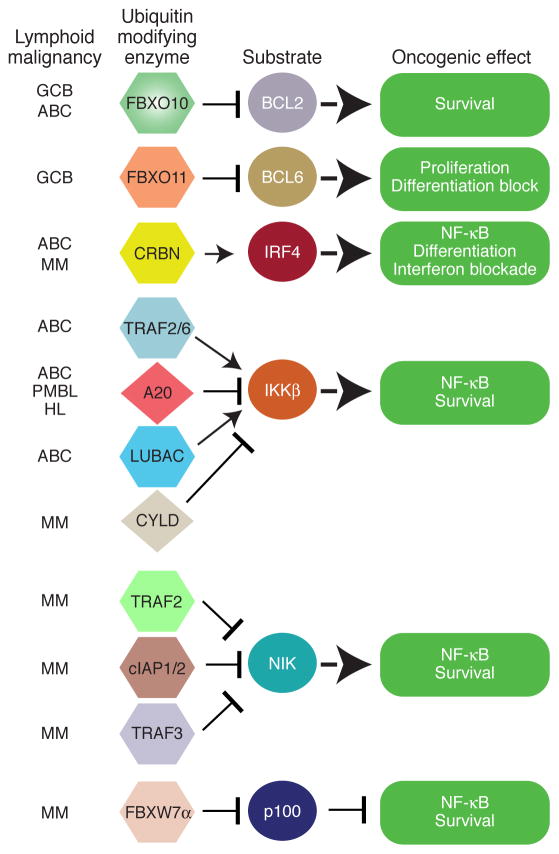
We review the central role of protein ubiquitination in the pathogenesis of lymphoid malignancies. In some cases, ubiquitin regulators are directly affected by oncogenic lesions, but more often they are required regulators of pathways that control proliferation, survival, and cell identity in these cancers (Table 1).
Table 1.
Overview of ubiquitination pathway components function in lymphoid malignancies.
| Ubiquitination pathway subunit | Ubiquitin enzyme complex | Gene symbol | Biochemical function | Biological function | Mutation status | Lymphoid malignancy | Reference |
|---|---|---|---|---|---|---|---|
| E2 conjugating enzyme | |||||||
| UBC13 | UBC13/UEV1A | UBE2N | K63 polyubiquitin chain formation | Canonical NF-κB activation | Unknown | ABC DLBCL | (89) |
| UEV1A | UBC13/UEV1A | UBE2V1 | K63 polyubiquitin chain formation | Canonical NF-κB activation | Unknown | ABC DLBCL | (89) |
| E3 ligase | |||||||
| RNF31 (HOIP) | LUBAC | RNF31 | IKKγ(NEMO) linear polyubiquitination | Canonical NF-κB activation | Gain-of-function SNPs | ABC DLBCL | (61, 62) |
| RBCK1 (HOIL-1L) | LUBAC | RBCK1 | IKKγ(NEMO) linear polyubiquitination | Canonical NF-κB activation | Unknown | ABC DLBCL | (61, 62) |
| SHARPIN | LUBAC | SHARPIN | IKKγ(NEMO) linear polyubiquitination | Canonical NF-κB activation | Unknown | ABC DLBCL | (61, 62) |
| TRAF2 | TRAF2 | K63 polyubiquitination | Canonical NF-κB activation | Missense mutation | ABC DLBCL | (34, 39) | |
| NIK stability | Non-canonical NF-κB inhibition | Inactivating mutation | Multiple myeloma | (83) | |||
| TRAF3 | TRAF3 | NIK stability | Non-canonical NF-κB inhibition | Inactivating mutation | Multiple myeloma | (82, 83) | |
| TRAF6 | TRAF6 | MALT1 K63 polyubiquitination | Canonical NF-κB activation | Unknown | ABC DLBCL# | (39, 40) | |
| c-IAP1 | BIRC2 | NIK stability | Non-canonical NF-κB inhibition | Inactivating mutation | Multiple myeloma | (82, 83) | |
| c-IAP2 | BIRC3 | NIK stability | Non-canonical NF-κB inhibition | Inactivating mutation | Multiple myeloma | (82, 83) | |
| FBXO10 | SCFFBXO10 | FBXO10 | BCL2 stability | Tumor suppressor | Deletion, missense mutation | GCB DLBCL; ABC DLBCL | (98) |
| FBXO11 | SCFFBXO11 | FBXO11 | BCL6 stability | Tumor suppressor | Deletion, missense mutation | GCB DLBCL | (99) |
| FBXW7 | SCFFBXW7 | FBXW7 | Nuclear p100 degradation | Non-canonical NF-κB activation | Unknown | Multiple myeloma | (118) |
| FBXW1A | SCFBTRC | BTRC | IκBα degradation | Canonical NF-κB inhibition | Unknown | Multiple myeloma, ABC DLBCL | (94–96) |
| Cereblon | CRL4CRBN | CRBN | IKZF1/3 degradation when bound to IMiD drug; decreased IRF4 | Survival | Unknown | Multiple myeloma, ABC DLBCL | (132–134) |
| Deubiquitinating enzyme | |||||||
| A20 | TNFAIP3 | Removing K63-linked ubiquitin chains; binding ubiquitin chains | Canonical NF-κB inhibition | Inactivating mutation | Hodgkin lymphomas; ABC DLBCL; MALT lymphoma | (34, 35, 70–72) | |
| CYLD | CYLD | Removing K63-linked ubiquitin chains | Canonical NFκB inhibition | Inactivating mutation | Multiple myeloma | (82, 83) |
UBE2N (UBC13), ubiquitin-conjugating enzyme E2N; UBE2V1 (UEV1A), Ubiquitin-Conjugating Enzyme E2 Variant 1; RNF31, ring finger protein 31; RBCK1, RanBP-type and C3HC4-type zinc finger containing 1; SHARPIN, SHANK-associated RH domain interactor; TRAF2, TNF receptor-associated factor 2; TRAF3, TNF receptor-associated factor 3; TRAF6, TNF receptor-associated factor 6; BIRC2 (cIAP1), baculoviral IAP repeat containing 2; BIRC3 (cIAP2), baculoviral IAP repeat containing 3; FBXO10, F-box protein 10; FBXO11, F-box protein 11; FBXW7, F-box and WD repeat domain containing 7; BTRC (FBW1A), beta-transducin repeat containing E3 ubiquitin protein ligase; CRBN, cereblon; CYLD, cylindromatosis (turban tumor syndrome); TNFAIP3 (A20), tumor necrosis factor alpha-induced protein 3; LUBAC, linear ubiquitin chain assembly complex; SCF, Skp, Cullin, F-box containing complex; CRL4, Cullin4–RING ligases; MALT, mucosa associated lymphoid tissue lymphoma translocation gene 1; MAP3K14 (NIK), mitogen-activated protein kinase kinase kinase 14; IKZF1, IKAROS family zinc finger 1; IKZF3, IKAROS family zinc finger 3; GCB DLBCL, germinal center B cell–like subtype of diffuse large B-cell lymphoma; ABC DLBCL, activated B-cell-like subtype of diffuse large B-cell lymphoma;
Although data are shown in lymphocytes, they are not supportive data in this cancerous phenotype.
Non-proteolytic protein ubiquitination in chronic active BCR signaling
Evidence that protein polyubiquitination is important in the oncogenic signaling of ABC DLBCL tumors came with the discovery that the DUB, A20, is frequently inactivated by various genetic and epigenetic means in ABC DLBCL tumors (34, 35). Specific deletion of TNFAIP3, encoding A20, in mouse B cells increases their responsiveness to multiple stimuli, including BCR crosslinking (36). As a consequence, they more readily generate both germinal center B cells and plasma cells, leading to autoimmunity as they age. A20 is recruited to the CBM complex in normal lymphocytes following antigen receptor signaling. This recruitment most likely relies upon the ability of A20 to bind polyubiquitin chains (see below). CBM assembly promotes polymerization of K63-linked ubiquitin chains and subsequent activation of TAK1 to phosphorylate IKKβ and stimulate NF-κB transcriptional activity (22). Once recruited, A20 curtails NF-κB activation, although the mechanism is complex and likely involves both enzymatic and non-enzymatic activities (37).
K63-linked ubiquitination in the CBM complex apparently involves members of the TRAF-family RING domain E3 ligase proteins. TRAFs are multidomain proteins that contain an N-terminal RING domain (with the exception of TRAF1), followed by a series of zinc finger domains and the TRAF domain, which is required for protein-protein interactions (38). MALT1 has a TRAF6 binding motif that it uses to recruit TRAF6 to the CBM complex and trigger TRAF6 auto-K63-linked polyubiquitination. In T cells, TRAF6 depletion almost completely abolishes IKK activation downstream of T-cell receptor signaling (39). Subsequent investigations revealed that TRAF6 functions in vitro and in vivo as an E3 ligase for MALT1, catalyzing its K63-linked ubiquitination (40). MALT1 lysine mutants that lack the ability to be modified by K63-linked ubiquitin chains are impaired in rescuing NF-κB signaling in MALT1 deficient cells (40), implying an essential function of MALT1 K63-linked ubiquitination. The other TRAF-family RING domain E3 ligase, TRAF2, can also bind MALT1 and mediate NF-κB activation during TCR signaling (39). Low frequency TRAF2 missense mutation (3%) was found in DLBCL, and one ABC-DLBCL-derived TRAF2 mutant was suggested to enhance NF-κB activity (34). The exact role of TRAF2 in CBM function will require further investigation. Moreover, given the fact that TRAF proteins have the potential to recruit other K63-linked ubiquitin ligases such as cIAP1/2 and Pellino, it cannot be ruled out that additional E3 ligases participate in CBM complex signaling. In addition to MALT1, BCL10 is modified by K63-linked polyubiquitination in the CBM complex during T-cell activation, although the responsible E3 ligase remains unknown (41). The BCL10 polyubiquitin chains interact with the regulatory γ subunit of IKK (IKKγ/NEMO), thereby recruiting IKK to the CBM complex and activating NF-κB.
MALT1 monoubiquitination and protease activity
In addition to acting as a scaffold protein within the CBM complex, MALT1 also contains a proteolytic activity that is constitutively activated in ABC DLBCL (42, 43). The defined MALT1 protease substrates include A20, BCL10, and RelB (44–46). Thus, MALT1 protease activity enhances NF-κB responses by inactivating A20 and biases the response away from the non-canonical NF-κB pathway by inactivating RelB. The specific MALT1 inhibitor z-VRPR-fmk inhibits NF-κB target gene expression and ABC DLBCL viability, making MALT1 an attractive therapeutic target (42, 43).
Recent studies revealed that the protease function of MALT1 is regulated by its monoubiquitination (47, 48). MALT1 is monoubiquitinated on Lysine 644, which is required and sufficient to activate its protease activity. This monoubiquitination promotes dimerization of MALT1, which is essential for its enzymatic activity. In ABC DLBCL cells, MALT1 is constitutively monoubiquitinated, and expression of a MALT1 mutant (K644R) that cannot be ubiquitinated is toxic to ABC DLBCL tumor cells. The E3 ubiquitin ligase required for MALT1 monoubiquitination in ABC DLBCL is currently unknown and deserves further investigation since inhibitors of this ligase activity could provide a therapeutic opportunity in ABC DLBCL.
Non-proteolytic protein ubiquitination in MYD88-IRAK signaling
Oncogenic MyD88 signaling and chronic active BCR signaling in ABC DLBCL activate NF-κB in a parallel and non-redundant fashion. The role of K63-linked polyubiquitination in MyD88-mediated innate immune response has been well documented (49). In Toll-like receptor (TLR) and IL-1 receptor signaling, MyD88 recruitment is mediated by heteromeric interactions between the MyD88 TIR domain and the TIR domains of the receptors. Receptor recruitment facilitates the interaction of MyD88 with IRAK4 and IRAK1, creating a complex in which IRAK4 phosphorylates IRAK1, leading to TRAF6 recruitment. TRAF6 then undergoes K63-linked auto-polyubiquitination, creating docking sites for proteins containing ubiquitin-binding domains. The NZF (Npl14 zinc finger) domain present in the TAK1 binding partner TAB2/3 displays K63-polyubiquitin binding preference (50). Recruitment of TAK1 to K63-polyubiquitinated TRAF6 leads to activation of TAK1 that in turn phosphorylates IKKβ (51, 52). IRAK1 is also modified with K63-linked ubiquitin chains during TLR signaling (53–55). Mutation of a lysine in IRAK1 that becomes polyubiquitinated impairs IL-1R/TLR4 induced NF-κB activation (55). Both TRAF6 and PELI1 (pellino E3 ubiquitin protein ligase 1) have been suggested to function as E3 ligases for IRAK1 during IL-1 receptor signaling (53, 55); however, the specific E3 ligase responsible for IRAK1 ubiquitination in TLR signaling remains unknown. In ABC DLBCL cells, direct evidence of K63-linked ubiquitination in MYD88-IRAK1-IRAK4 signaling is missing. However, interference with the MyD88 pathway in ABC DLBCL reduces TAK1 phosphorylation demonstrably (authors’ unpublished data), suggesting a role for TRAF6 ubiquitination in this oncogenic signaling. Going forward, it will be important to identify the ubiquitination status of IRAK1 and TRAF6 in the constitutive MyD88 signalosomes in ABC DLBCL.
Linear ubiquitination in ABC DLBCL
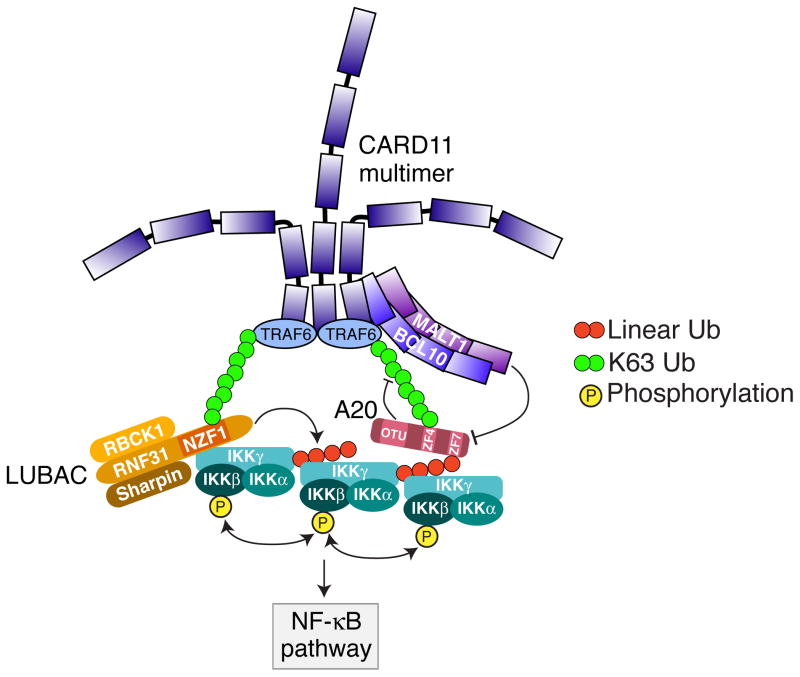
Protein modification with linear ubiquitin chains plays a selective role in signaling pathways leading to NF-κB activation (31–33, 56). The linear ubiquitin chain assembly complex (LUBAC), composed of RNF31 (HOIP), RBCK1 (HOIL-1L), and SHARPIN, is the only known E3 ligase capable of assembling linear polyubiquitin chains. RNF31 and RBCK1 belong to the RBR class of E3 ligases, but only the RBR domain of RNF31 is required for linear ubiquitin chain formation by LUBAC (24, 57). However, a combination of RNF31 with either RBCK1 or SHARPIN is the minimal requirement for linear ubiquitin chain catalysis (24, 31). In the classical NF-κB pathway, LUBAC specifically recognizes and targets IKKγ/NEMO for linear polyubiquitination, which is required for IKK activation (56, 58). The LUBAC subunit RNF31 contains an NZF domain that directly interacts with IKKγ (59) (Fig. 1). This same domain uses different amino acids to bind to K63-linked polyubiquitin chains, which allows LUBAC to be recruited to receptor complexes containing TRAFs or other K63-specific ubiquitin ligases. The linear ubiquitination of IKKγ could provide a mechanism for recruitment of additional IKK complexes to the signalosome since the UBAN domain of IKKγ binds to linear ubiquitin chains, using residues that are distinct from those recognized by LUBAC (59, 60). The proximity of multiple IKK complexes could in principle result in autophosphorylation, thereby increasing NF-κB pathway activation (59) (Fig. 1).
Fig. 1. LUBAC mediated NF-κB activation in ABC DLBCL.
The LUBAC subunit RNF31 contains an NZF domain that binds to K63-linked polyubiquitin chains, which allows LUBAC to be recruited to the CARD11-BCL10-MALT1 (CBM) signalosome complex containing TRAF6 or other K63-specific ubiquitin ligases. Simultaneously, this same domain directly interacts with IKKγ and leads to IKKγ linear ubiquitination. The linear ubiquitinated IKKγ recruits additional IKK complex through its UBAN domain which binds to linear ubiquitin chains. The proximity of multiple IKK complexes results in autophosphorylation and activation of IKKβ. A20, a negative regulator in NF-κB pathway, can bind to both K63-linked and linear ubiquitin chains through the c-terminal zinc finger #4 (ZF4) and zinc finger #7 (ZF7) respectively. The linear ubiquitination of IKKγ thus recruits A20 to the CBM complex and promotes MALT1 cleavage of A20. Both mechanisms result in increasing NF-κB pathway activation in ABC DLBCL.
LUBAC plays an essential role in the oncogenic activation of NF-κB in ABC DLBCL cells since depletion of LUBAC subunits by RNAi reduces constitutive NF-κB activity and is lethal to ABC DLBCL lines (61, 62). A major substrate of LUBAC in ABC DLBCL cells is IKKγ, and LUBAC is essential for optimal IKK activity in these cells (61). LUBAC associates with the CBM complex in ABC DLBCL cells and stimulates MALT1 protease activity, showing that LUBAC plays a direct role in chronic active BCR signaling (61, 62). LUBAC may also contribute to normal BCR signaling, since knockdown of RNF31 impaired IKK activation following crosslinking of the BCR in a GCB DLBCL cell line (61, 62). Conditional deletion of the LUBAC subunit RNF31 in normal mouse B cells severely impairs T-cell-dependent and –independent antibody responses (63). Experiments with RNF31-deficient B cells did not show an impairment of NF-κB activation following BCR crosslinking (63), which is at odds with findings in lymphoma cell lines. This apparent discrepancy could be explained by differences in the degree of BCR cross-linking in the two studies and/or in the differentiation stage of B cells analyzed.
How LUBAC is recruited to the CBM complex is unclear at this stage. However, recruitment of LUBAC to receptor complexes in other signaling pathways is dependent on polyubiquitin modification of receptor-associated factors (58). Thus, it is likely that LUBAC binds to the CBM complex through polyubiquitin chains attached to one or more components in this complex. As discussed previously, multiple CBM subunits are modified by ubiquitin following receptor stimulation (39, 40, 47). Further work is needed to determine the ubiquitination status CBM complex components in ABC DLBCL and to identify which modifications are required for LUBAC recruitment and IKKγ linear ubiquitination.
Genetic analysis of human lymphomas identified rare germ line polymorphisms (SNPs) in RNF31 that increase LUBAC complex formation and activity (61). Human RNF31 contains an amino-terminal zinc finger domain required for NEMO binding, followed by a ubiquitin-associated domain (UBA) and a carboxy-terminal RBR E3 ligase domain (24). The RNF31 UBA domain interacts with the ubiquitin-like (UBL) domain of RBCK1, thereby promoting LUBAC enzymatic activity (56, 64). A small region of the RNF31 UBA domain, from amino acids 579 to 623, mediates binding to the UBL domain of RBCK1 (64), and this is the region that is affected by the lymphoma-associated RNF31 SNPs. These two SNPs – Q584H and Q622L – are rare among healthy individuals (combined prevalence ~1%) but are highly enriched among patients with ABC DLBCL (7.8%) (61). The amino acid substitutions caused by the RNF31 SNPs increase the interaction of RNF31 and RBCK1 and consequently increase LUBAC ubiquitin ligase activity and downstream NF-κB signaling (61). Based on studies of the Jurkat T-cell line, it has been suggested that LUBAC may activate NF-κB in a manner that is independent of its catalytic activity (62). However, the genetic and functional data in ABC DLBCL suggest that the ubiquitination of IKKγ by LUBAC is critical for NF-κB activity and cell survival (61).
LUBAC potentially could serve as a therapeutic target in ABC DLBCL. A peptide representing the α-helix of RNF31 that contacts RBCK1, with the Q622L SNP-associated amino acid substitution, was able to inhibit LUBAC complex formation in ABC DLBCL cells, decrease NF-κB activity and kill these cancer cells. This suggests that a small molecule screen for inhibitors of the RNF31-RBCK1 interaction might be a worthwhile endeavor. Inherited complete RBCK1 deficiency is associated with severe inflammation and bacterial infections in children (65). It is nonetheless possible that partial inhibition of LUBAC function in adults could have manageable side effects while controlling the growth of tumors that depend on this enzyme.
Deubiquitinating enzymes in lymphoid malignancies
Like protein phosphorylation, ubiquitination is a reversible process mediated by specific proteases named DUBs. Roughly 100 DUBs are encoded in the human genome (66), implying that DUBs regulate diverse biological processes by removing ubiquitin from many different substrates. Two DUBs that regulate NF-κB signaling – A20 and CYLD – are targeted by recurrent genetic alterations in human lymphoid malignancies.
A20 in lymphoid malignancies
As mentioned above, the gene encoding A20, TNFAIP3, is targeted by a variety of recurrent mutations, deletions, and translocations, or is epigenetically silenced in lymphoid malignancies, resulting in partial or total loss of A20 function. That A20 would function as a tumor suppressor in these cancers is not surprising given the fact that TNFAIP3 is an NF-κB target gene whose induction provides negative feedback that limits the output of this pathway.
A20 contains two domains with distinct functions: an amino-terminal ovarian tumor (OTU) domain with deubiquitinating activity and a carboxy-terminal domain consisting of seven zinc fingers that contributes to its ubiquitin ligase activity and to its ability to bind polyubiquitin chains (Fig. 1). The way in which these various domains and activities of A20 inhibit NF-κB signaling is an area of active and evolving research. One model posits that A20 plays a ubiquitin ‘editing’ role by removing K63-linked ubiquitin chains while also attaching K48-linked polyubiquitin chains, leading to substrate degradation (67). However, recent experiments have shown that the ability of A20 to bind ubiquitin chains using its zinc finger domain is necessary and sufficient to block IKK and NF-κB activation, while suggesting that its E3 enzymatic activity is at least partially dispensable (68, 69). Thus, A20 interacts with and modifies ubiquitinated protein substrates in multiple ways, perhaps using a variety of biochemical mechanisms to regulate NF-κB responses.
A20 is frequently inactivated in B-cell lymphomas: ~45 % of classical Hodgkin’s lymphomas, 25 % of ABC DLBCLs, and 20 % of MALT lymphomas harbor inactivating mutations of at least one allele of TNFAIP3 (34, 35, 70–72). In addition, A20 is a protease substrate of MALT1 and can be cleaved in tumors in which MALT1 is activated (44). Reintroduction of A20 into A20-null ABC DLBCL cell lines results in growth arrest and cell death (34, 35), supporting its tumor suppressor role.
Despite this genetic evidence, the direct endogenous targets of A20 in lymphomas have not been conclusively identified. In ABC DLBCL tumors, inactivation of A20 can co-occur with both CD79B and MYD88 mutations, suggesting that A20 could be involved in chronic active BCR signaling and in oncogenic MYD88 signaling (19). In normal immune cells, the identified substrates of A20 deubiquitinating enzyme activity include TRAF6, MALT1, and IKKγ (67, 73, 74), and these proteins are likely to be modified with K63-linked polyubiquitin in lymphomas as well.
Recent reports suggest that non-catalytic functions of A20 are key to its ability to terminate NF-κB responses. Zinc finger #4 (ZF4) of A20 binds K63-linked polyubiquitin chains and this binding contributes to its ability to inhibit NF-κB activation in response to TNFα (68, 69). Recently, zinc finger #7 (ZF7) of A20 has been shown to specifically bind linear ubiquitin chains and has no affinity for other ubiquitin linkages (75, 76). A20 utilizes both ZF4 and ZF7 to suppress TNFα-mediated NF-κB activation (75, 76) (Fig. 1). Interestingly, some lymphoma-associated missense mutations target A20 ZF7, creating isoforms that cannot bind linear ubiquitin chains (75). In ABC DLBCL, LUBAC activity promotes linear ubiquitination of IKKγ, which recruits A20 to the CBM complex. Moreover, A20 recruitment to CBM complex is increased by expression of RNF31 SNP-associated gain-of-function isoforms, most likely because of their enhancement of LUBAC activity (61). Of note, MALT1 cleavage of A20 is increased by its recruitment to the CBM, potentially due to the increased proximity of A20 to MALT1. The relative influence of linear and K63-linked polyubiquitin chains on A20 recruitment in lymphomas deserves further study.
CYLD in lymphoid malignancies
The largest subfamily of DUBs, the ubiquitin-specific proteases (USPs), consists of more than 50 members in humans. Although the function of most USPs has yet to be characterized, one member, cylindromatosis (CYLD), has been extensively studied as a classic tumor suppressor (77). CYLD was originally identified as a gene associated with familial cylindromatosis (FC), a neoplasm of the hair follicle stem cell. FC patients carry one loss-of-function CYLD allele in their germ line and the tumors that arise acquire a second inactive CYLD allele. The inactivating genetic events target exons encoding the catalytic domain, implying that the DUB activity of CYLD is critical for its tumor suppressor function. Subsequent studies revealed that CYLD acts as a tumor suppressor by negatively regulating the NF-κB pathway (78–80). CYLD selectively removes K63-linked polyubiquitin chains, and its substrates include IKKγ/NEMO, TRAF2, TRAF6, TRAF7, RIP1, and TAK1 (77). Thus, CYLD could function as a tumor suppressor in B-cell malignancies in which classical NF-κB pathways are constitutively activated to promote tumor cell survival. However, CYLD is a protease substrate of MALT1 (81), suggesting that oncogenic activation of the CBM complex might obviate the need for genetic inactivation of CYLD. Indeed, CYLD mutations do not occur at high frequency in ABC DLBCL.
CYLD inactivation is a recurrent but relatively low frequency (1–2%) event in multiple myeloma (82, 83). In this malignancy, NF-κB is activated due to a variety of genetic lesions that lead to stable, high expression of the kinase NIK. NIK can directly phosphorylate IKKβ, activating the classical NF-κB pathway, as well as IKKα, activating the alternative NF-κB pathway. In multiple myeloma cell lines, inhibition of IKKβ with a specific small molecule inhibitor is lethal, suggesting an important role for the classical NF-κB pathway. The functionally relevant targets of CYLD in multiple myeloma are currently unknown, and may yield further insight into the mechanism of NF-κB activation in this malignancy.
Ubc13-containing E2 enzymes in ABC DLBCL
Ubc13 (UBE2N) is the catalytic subunit of E2 enzymes that attach K63-linked polyubiquitin chains to their substrates (84). UBC13 forms an enzyme complex with either Uev1A (UBE2V1) or Mms2 (UBEV2). In response to various stimuli that activate NF-κB, membrane receptors recruit the Ubc13/Uev1A complex, which collaborates with an E3 ligase (e.g. TRAF6 or TRAF2) to catalyze the formation of K63-linked polyubiquitin chains, leading to TAK1 and IKK activation (85). In DNA replication and repair pathways, the Ubc13/Mms2 complex promotes K63-linked ubiquitination of PCNA, Rad5, and Pol30, leading to the recruitment of repair proteins to DNA lesions (86–88).
An essential oncogenic role of Ubc13/Uev1A in ABC DLBCL has recently been suggested (89). A small molecule (NSC697923) targeting the E2 enzyme activity of the Ubc13/Uev1A complex was able to reduce NF-κB activity in ABC DLBCL cells, likely by inhibiting K63 polyubiquitin-dependent IKK activation. Consequently, significant growth inhibition and apoptosis was observed in NSC697923-treated ABC DLBCL cells. UBC13 depletion by RNAi also reduced NF-κB activation and proliferation/survival of ABC DLBCL cells. These findings provide evidence that K63-mediated polyubiquitin chains regulate NF-κB signaling in ABC DLBCL, suggesting the potential involvement of TRAF family proteins. Further investigations are needed to elucidate the specific targets of Ubc13/Uev1A in ABC DLBCL and to identify the E3 ligases that collaborate with this E2 enzyme in these cancer cells. These findings also raise the possibility that Ubc13/Uev1A could serve as a therapeutic target in ABC DLBCL. However, Ubc13/Uev1A inhibition was also toxic to GCB DLBCL cell lines that do not rely on NF-κB, implying that other cellular processes required for cellular viability also utilize this E2 enzyme (89). Given the many pathways that may be involved, it will be important to undertake animal studies to determine whether a therapeutic window might exist for Ubc13/Uev1A inhibitors.
SCF E3 ligase complexes in lymphoid malignancies
Many RING-E3 ligases form multi-protein complexes in which other subunits provide substrate binding specificity. This is exemplified by the SCF (Skp1-Cul1-F-box) complex E3 ligases, which attach K48-linked polyubiquitin to their substrates leading to proteasomal degradation. SCF E3 ligase complexes generally include three static subunits: the catalytic RING subunit RING-box protein 1 (RBX1), a scaffolding Cullin (Cul1) subunit, and S-phase kinase associated protein 1 (Skp1). In addition, these ligases contain one variable subunit belonging to the F-box protein family, which serves as the substrate recruitment module and determines the target specificity of the ligase. The 69 F-box proteins that are encoded in human genome can be classified into three subfamilies based on their carboxy-terminal protein binding domains: the FBXW family containing WD40 repeat domains, the FBXL family containing leucine-rich repeat domain, and the FBXO family containing other domains (90). SCF family E3 ligase complexes typically require a post-translational modification of their substrate for efficient recognition. In most cases, phosphorylation at specific ‘phospho-degron’ motifs in the substrate is necessary of optimal ubiquitination (91). The collaboration of a protein kinase(s) with an E3 ligase provides an additional layer of regulation aimed at achieving appropriate protein levels and regulating protein half-lives differentially under various physiological conditions.
The F-Box protein E3 ligases play critical roles in multiple cellular processes by ubiquitinating key regulatory proteins in these processes. For example, the F-box protein FBXW1A (also called β-TRCP) targets IκBα, a major inhibitor of the classical NF-κB pathway, in a well-characterized phosphorylation-dependent manner (92, 93). In human B-cell malignancies, enhanced FBXW1A expression was observed in melanoma cell lines (94, 95). Hence inhibitors of the ubiquitin ligase containing β-TRCP would be a potential therapeutic strategy in ABC DLBCL and other cancers that rely upon the classical NF-κB pathway. One strategy to inhibit SCF ubiquitin ligases is to target the required modification of the Cullin subunit by the short peptide NEDD8. MLN4924, a small molecule inhibitor of the NEDD8 activating enzyme (NAE), effectively blocks NF-κB activity in ABC DLBCL cells and is effective in treating ABC DLBCL xenografts (96). Of course, SCF ubiquitin ligases perform many other cellular functions including regulation of the cell cycle, and therefore NAE inhibitors that will have other on-target effects besides inhibiting NF-κB that may limit their clinical utility.
Dysregulation of F-box protein function is key to the pathogenesis of many human malignancies (97). We review recent genetic and functional evidence demonstrating their importance in lymphoid cancers.
FBXO10 and FBXO11
Two FBXO family members, FBXO10 and FBXO11, have been reported recently to be essential regulators in DLBCL (98, 99). Both proteins are highly homologous to a C. elegans protein dre-1 that interacts and regulates the C. elegans homologue of BCL-2, CED-9, thereby influencing cell death in the worm (98).
FBXO10 contains one conserved F-box domain in the amino-terminal region followed by a substrate recognition domain consisting of 17 parallel β-helix (PbH1) repeats and CASH (carboxyl-terminal carbohydrate binding proteins and sugar hydrolases) domains. In DLBCL, the anti-apoptotic oncoprotein BCL2 has been demonstrated to be a primary substrate of SCFFBXO10 (98). FBXO10 directly binds to BCL2, targets BCL2 for ubiquitination and promotes its degradation, thereby regulating BCL2 protein levels in human lymphoma cells. Inducible expression of FBXO10 in human lymphoma lines is toxic due to increased apoptosis, and this toxicity can be partially rescued by ectopic provision of BCL2, identifying BCL2 as a key SCFFBXO10 substrate in these cancer cells.
Several lines of evidence suggest that FBXO10 functions as tumor suppressor in human lymphomas. Compared with normal germinal center B cells, a majority of GCB DLBCL tumors have much lower expression of FBXO10 (98). FBXO10 is also deleted or mutated in DLBCLs as less frequent mechanisms that limit its function. FBXO10 coding region mutations were detected in 5% of GCB DLBCLs and in 1% of ABC DLBCLs, including both frameshift and missense mutations. One missense mutation (R44H) is located to the F-box domain and prevents the association of FBXO10 with the SCF complex. Two others (V762L and R825W) are located in the CASH domains and presumably influence substrate recruitment and/or ubiquitination, although the exact mechanisms have not been elucidated. In functional studies, all three FBXO10 missense mutants have reduced ability to destabilize BCL2 protein and are much less toxic to lymphoma cells than wildtype FBXO10, consistent with a loss-of-function mechanism. All FBXO10 mutations that have been described in DLBCL tumors are heterozygous, implying that complete loss of FBXO10 is not compatible with lymphoma development and that FBXO10 functions as a haploinsufficient tumor suppressor.
FBXO11 also functions as a tumor suppressor in DLBCL, albeit by an entirely different mechanism than FBXO10 (99). FBXO11 contains an amino-terminal F-box domain and several CASH domains that presumably function in substrate recognition. Unlike FBXO10, which resides primarily in the cytoplasm (98), FBXO11 is confined to the nucleus where it binds and ubiquitinates the transcription factor BCL6, leading to its degradation (99). Translocations and other genetic events frequently dysregulate BCL6 expression in DLBCL, blocking terminal plasmacytic differentiation, inhibiting apoptosis and DNA damage responses, and promoting cell cycle progression, among many other phenotypes (100).
Ectopic expression of FBXO11 in DLBCL lines that lack endogenous FBXO11 expression inhibits proliferation and induces apoptosis, consistent with a tumor suppressor function. Indeed, FBXO11 is deleted in ~15% of DLBCL cell lines and mutated in ~4% of primary DLBCL tumor biopsies (99). DLBCL samples harboring FBXO11 mutations have elevated BCL6 levels consistent with BCL6 being a relevant target of FBXO11 in DLBCL. The FBXO11 mutants are impaired in their ability to localize in the nucleus and bind BCL6 and \are unable to degrade BCL6 efficiently (99), supporting the hypothesis that the tumor suppressor function of FBXO11 in DLBCL is related to its ability to target BCL6 for degradation.
A remaining interesting puzzle is the identity of the kinases that are required in DLBCL to phosphorylate the substrates of SCFFBXO10 and SCFFBXO11. The turnover rates of both BCL2 and BCL6 are phosphorylation-dependent (101–105). The kinase ERK can phosphorylate BCL6 upon BCR activation in human B cells, causing its degradation (101). However, the recognition of BCL6 by FBXO11 seems to be ERK-independent and does not rely on BCR activation in DLBCL (106). Multiple kinases, including JNK, CDC2, RAF-1, mTOR, AKT, and CDK6, can phosphorylate BCL2 in response to multiple stimuli (106). Identification of the signaling pathways that regulate the phosphorylation of the FBXO10 and FBXO11 substrates in DLBCL is an important goal. In particular, pharmacological inhibition of phosphatases that act on the degrons of BCL2 and BCL6 could serve as drug targets in DLBCL.
FBXW7
FBXW7, a member of the FBXW F-Box protein family, has eight WD-40 repeats in its carboxy-terminal region. Structurally, these WD-40 repeats form a β-propeller binding pocket that utilizes three conserved arginine residues to form contacts with phosphorylated targets (107, 108). SCFFBXW7 targets multiple substrates, including cyclin E, c-MYC, c-JUN, MCL-1 and NOTCH1, that are important oncogenes involved in tumor proliferation, growth, and survival (109, 110). FBXW7 is mutated in many cancer types, and among lymphoid malignancies it plays a prominent role in T-cell acute lymphoblastic leukemia (T-ALL), which acquire FBXW7 mutations in 20–30% of cases (109, 111). The FBXW7 mutations in T-ALL are predominantly heterozygous and cluster within the WD40 domains of the protein. The three highly conserved arginine residues that directly interact with phosphorylated substrates are most frequently mutated and account for around 40% of all tumor-derived mutations in FBXW7 (112). These FBXW7 mutations in T-ALL affect the stability of the transcription factors NOTCH1 and c-MYC (113, 114). Indeed, T-ALLs acquire NOTCH1 mutations affecting the PEST domain, resulting deletion of a conserved degron motif that is recognized by FBXW7 (115). In a knockin animal model carrying the most frequent T-ALL FBXW7 mutation, the proportion of leukemia-initiating cells is increased and c-MYC is stabilized (116). Of note, full genetic inactivation of FBXW7 by mutation or deletion is relatively rare in T-ALL (114, 117), implying that the recurrent FBXW7 missense mutants may not be ‘dead’ but instead may selectively alter the substrates and/or functions of the protein.
In contrast to the frequent FBXW7 mutations in T-ALL, alterations of the FBXW7 gene are rare in B-cell malignances, suggesting that FBXW7 might have a different function in these cancers. Indeed, a recent study identified a potentially oncogenic role in multiple myeloma for FBXW7α, which is the alternatively spliced isoform of FBXW7 that resides predominantly in the nucleus (118). An important substrate of FBXW7α is the NF-κB family member p100, which binds and inhibits preformed NF-κB heterodimers (p50/RelA, p50/RelB) (119). When the alternative NF-κB signaling pathway is activated, IKKα phosphorylates p100 in the cytoplasm, leading to its degradation in an SCFβTRCP-dependent manner. By contrast, FBXW7α targets nuclear p100 for degradation in a glycogen synthase kinase 3 (GSK3)-dependent manner (118). As mentioned above, myelomas frequently activate both the classical and alternative NF-κB pathway (82, 83), and the constitutive degradation of p100 by FBXW7α is required to maintain NF-κB activity and viability of these cancer cells (118). Thus, the role of FBXW7 in cancer is context dependent, either performing a tumor suppressor function, as in T-ALL, or an essential, positive function, as in multiple myeloma, depending on which of its substrates contribute most to the malignant phenotype in each context.
Cereblon E3 ligase and the IMiD drugs
While the SCF family E3 ligases include Cullin1 (CUL1), some Cullin–RING ligases in the CRL4 subfamily use Cullin4 (CUL4) as the scaffolding subunit. CLR4 ligases incorporate either of two functionally redundant paralogues, CUL4A and CUL4B, and the catalytic RING subunit RBX1. In addition, these ubiquitin ligase complexes contain DNA damage-binding protein 1 (DDB1), a triple WD40 domain-containing protein that serves as an adaptor to recruit members of the DDB1–CUL4 associated factors (DCAF) family of substrate receptors (120–123). The critical role of CRL4 ligases in human hematologic malignancies has been revealed with the discovery that the small molecule therapeutics in the immunomodulatory (IMiD) class bind and alter the activity of Cereblon (CRBN), a DCAF for the CRL4 E3 ligase family (124).
Cereblon is encoded by the CRBN gene located in chromosomal region 3p26.2, which is associated with autosomal recessive nonsyndromic mental retardation (ARNSMR), potentially linking Cereblon to central nervous system development (125, 126). Indeed, an inherited nonsense Cereblon mutation (R419X) that truncates 24 amino acids in the carboxy-terminal region is associated with ARNSMR (127). Although the molecular mechanism of Cereblon function in ARNSMR remains unsolved, some studies indicate that the role of Cereblon may be to regulate the assembly and surface expression of ion channels and AMP-activated protein kinase (AMPK) function (128–131). The first clue linking Cereblon to ubiquitin ligase activity came from a search for proteins that are associated with the CUL4-DDB1 E3 ligase complex, in which Cereblon, along with 30 other proteins, was identified as a DCAF with a potential function in substrate recognition (120).
The IMiD drug class includes thalidomide, lenalidomide, and pomalidomide, which are used in the treatment of multiple myeloma and myelodysplastic syndromes and are currently being clinically evaluated in a variety of lymphoid malignancies, including DLBCL, CLL, and MCL. The great success of these drugs clinically preceded an understanding of their mechanism of action, which was first revealed by the discovery that thalidomide covalently binds to Cereblon in a protein complex containing DDB1 (124). Subsequent studies demonstrated that Cereblon is also a target for lenalidomide and pomalidomide in both multiple myeloma and ABC DLBCL (132–134). Depletion of Cereblon by RNAi makes ABC DLBCL cells insensitive to lenalidomide, providing genetic evidence that this E3 ligase is the target of lenalidomide in ABC DLBCL (133). In multiple myeloma, a cell line rendered resistant to lenalidomide was shown to have acquired a homozygous deletion of the gene encoding Cereblon, providing genetic evidence implicating this ubiquitin ligase in the mechanism of action of IMiDs in multiple myeloma (132).
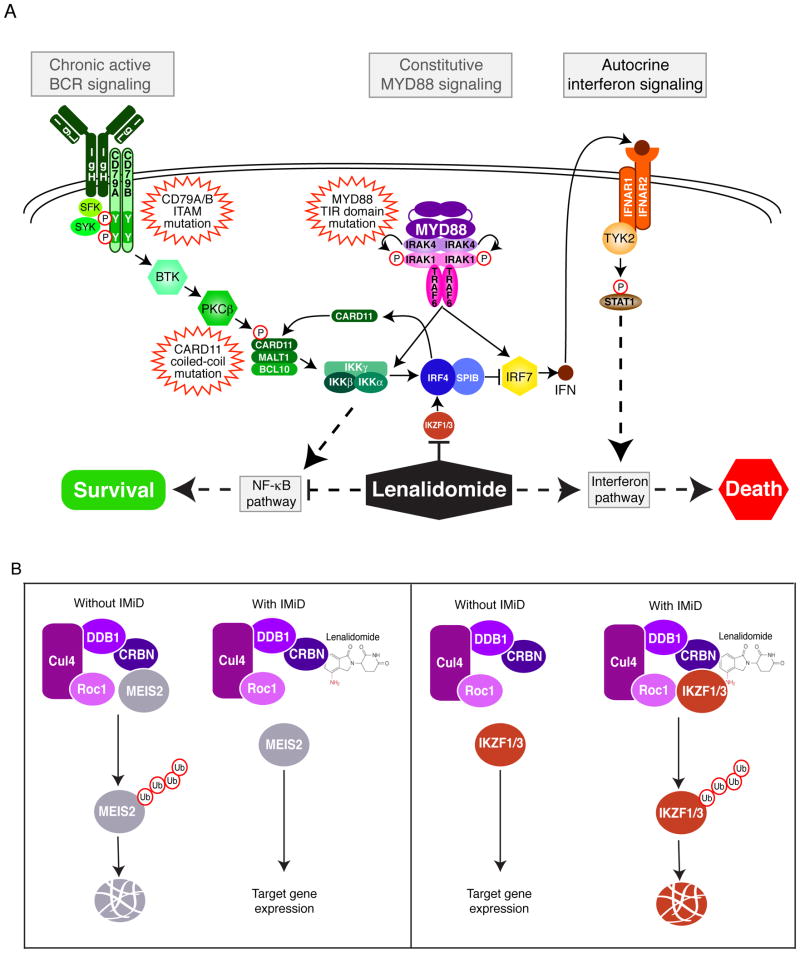
A principle consequence of IMiD modification of Cereblon in myeloma and ABC DLBCL is the downregulation of IRF4, a master transcriptional regulator in these cancers (132, 133). IRF4 was discovered to be essential for the survival of all multiple myeloma types, irrespective of their underlying genetic aberrations, apparently due its control of multiple target genes including MYC (135). IRF4 is also an essential transcription factor in ABC DLBCL, where it acts in concert with the transcription factor SPIB, which is also downregulated by lenalidomide treatment (Fig. 2A). The toxicity of lenalidomide for ABC DLBCL cells can be attenuated by ectopic expression IRF4, demonstrating that IRF4 plays a central role in the action of IMiDs in these lymphomas (133). IRF4 inhibition causes the death of ABC DLBCL cells by two complementary mechanisms. First, IRF4 represses IRF7, which encodes a key transcription factor controlling type I interferon production. Downmodulation of IRF4 expression by lenalidomide increases IRF7 expression, thereby enhancing synthesis and secretion of interferon β (IFNβ), which kills ABC DLBCL cells by autocrine signaling through the interferon receptor. The second way lenalidomide kills ABC DLBCL cells is by decreasing expression of CARD11, which is directly transactivated by IRF4, with a consequent decrease in NF-κB. The ability of ABC DLBCL to produce IFNβ in response to lenalidomide is predicated on oncogenic signaling by MYD88 whereas the effect of lenalidomide on NF-κB is a consequence of chronic active BCR signaling in these lymphomas. Thus, lenalidomide exploits the oncogenic abnormalities of ABC DLBCL, thus providing a measure of therapeutic specificity that allows it to be tolerated when administered as a drug.
Fig. 2. Function of lenalidomide and CRBN in ABC DLBCL.
(A). In ABC DLBCL, oncogenic mutations in both the BCR and MyD88 pathways provide pro-survival NF-κB signaling. However, MyD88 signaling also induces IFNβ which is harmful to ABC DLBCL survival. Lenalidomide targets IKZF1/3, which leads to the downregulation of IRF4 and SPIB expression. Downmodulation of IRF4/SPIB increases IRF7 expression, thereby enhancing synthesis and secretion of interferon β (IFNβ), and decreases CARD11 expression, thereby inhibiting NF-κB signaling. (B). Binding of IMiD compounds to CRBN can either prevent CRL4Cereblon from degrading its natural substrate (MEIS2) or modify the substrate specificity of CRL4Cereblon (IKZF1 and IKZF3), thus alters the impact of CRL4Cereblon on the proteome.
Attacking Cereblon with IMiD drugs represents an attractive therapeutic strategy in ABC DLBCL. Indeed, lenalidomide has shown preferential activity against ABC DLBCL in early phase clinical trials (136) and is now being evaluated in combination with chemotherapy in this lymphoma subtype. Since IRF4 is a direct NF-κB target gene, its expression can also be decreased by blocking chronic active BCR signaling with ibrutinib. Consequently, lenalidomide and ibrutinib synergize in killing ABC DLBCL cells in vitro and in xenograft models (133), prompting a currently active clinical trial combining these two drugs with chemotherapy in relapsed/refractory ABC DLBCL (http://clinicaltrials.gov/ct2/show/NCT02142049).
Treatment of ABC DLBCL or multiple myeloma cells with lenalidomide decreases IRF4 mRNA levels, suggesting that it acts through CRL4Cereblon to decrease the expression of a transcription factor or epigenetic regulator that positively regulates IRF4 transcription. Two B-cell-specific transcription factors, Ikaros family zinc finger proteins 1 and 3 (IKZF1 and IKZF3), bind to CRL4Cereblon in the presence of lenalidomide, leading to their ubiquitination and degradation (137–139). The toxicity of lenalidomide is abolished in myeloma cells carrying an IKZF3 mutant isoform that is resistant to Cereblon-dependent degradation, indicating that IKZF3 degradation is necessary for lenalidomide’s action in these cells (138). Depletion of IKZF1 or IKZF3 decreases IRF4 transcription, suggesting that they may be direct IRF4 transactivators (138, 139).
A recent study based on the structure of the CRL4Cereblon E3 ubiquitin ligase complex identified the homeobox transcription factor MEIS2 as an endogenous substrate of Cereblon and showed that IMiD compounds block binding of MEIS2 to CRL4Cereblon, thereby preventing its degradation (140). Thus, IMiD compounds can modify the impact of CRL4Cereblon on the proteome by two distinct mechanisms, either by preventing CRL4Cereblon from degrading its natural substrates (as in the case of MEIS2) or by modifying the substrate specificity of CRL4Cereblon (as in the cases of IKZF1 and IKZF3) (Fig. 2B).
Conclusion and perspectives
The significance of protein ubiquitination in the pathogenesis of human lymphoid malignancies is indisputable (Fig. 3). Nonetheless, our understanding of the functions of E3 ligases, E2 conjugating enzymes and deubiquitinating enzymes in these cancers is still at an elementary stage. Most of our current models of how protein ubiquitination regulates the malignant behavior of lymphoid cancers are derived from studies in normal lymphocytes. Therefore, it will be important to study the ubiquitin modifying machinery in the context of cell line models of lymphoid malignancies, identifying co-factors that modify the activity and/or specificity of these enzymes.
Fig. 3. Protein ubiquitination in lymphoid malignancies pathogenesis pathways.
Showing is the overview the central role of protein ubiquitination in the pathogenesis of lymphoid malignancies. In some cases, ubiquitin regulators are directly affected by oncogenic lesions, but more often they are required regulators of pathways that control proliferation, differentiation, survival, and cell identity in these cancers. See main text for details.
The protein ubiquitination system is a promising therapeutic target in a variety of human lymphoid malignancies but our knowledge of how to target this machinery with small molecules is rudimentary. The ability of a small peptide to interfere with the assembly of the LUBAC ubiquitin ligase suggests that small molecules that disrupt protein-protein interactions important for the function of the ubiquitin machinery could be developed. The example of the IMiD drugs raises the possibility that ubiquitin ligase specificity can be modulated by small molecules that change the nature of the substrate recognition domain. Indeed, each of the IMiDs has quantitatively and qualitatively different biological effects, most likely due to the way in which they modify substrate specificity. We can thus be hopeful that small molecules can be designed to modify the proteome of cancer cells in ways that interfere with their proliferation or survival, taking advantage of the central role played by ubiquitination in regulating key biological processes.
Acknowledgments
This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
Footnotes
The authors have no conflicts of interest to declare.
References
- 1.Shaffer AL, 3rd, Young RM, Staudt LM. Pathogenesis of human B cell lymphomas. Annual review of immunology. 2012;30:565–610. doi: 10.1146/annurev-immunol-020711-075027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alizadeh AA, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 3.Wright G, Tan B, Rosenwald A, Hurt EH, Wiestner A, Staudt LM. A gene expression-based method to diagnose clinically distinct subgroups of diffuse large B cell lymphoma. Proc Natl Acad Sci U S A. 2003;100:9991–9996. doi: 10.1073/pnas.1732008100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dave SS, et al. Molecular diagnosis of Burkitt’s lymphoma. N Engl J Med. 2006;354:2431–2442. doi: 10.1056/NEJMoa055759. [DOI] [PubMed] [Google Scholar]
- 5.Savage KJ, et al. The molecular signature of mediastinal large B-cell lymphoma differs from that of other diffuse large B-cell lymphomas and shares features with classical Hodgkin lymphoma. Blood. 2003;102:3871–3879. doi: 10.1182/blood-2003-06-1841. [DOI] [PubMed] [Google Scholar]
- 6.Rosenwald A, et al. Molecular diagnosis of primary mediastinal B cell lymphoma identifies a clinically favorable subgroup of diffuse large B cell lymphoma related to Hodgkin lymphoma. J Exp Med. 2003;198:851–862. doi: 10.1084/jem.20031074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bergsagel PL, Chesi M, Nardini E, Brents LA, Kirby SL, Kuehl WM. Promiscuous translocations into immunoglobulin heavy chain switch regions in multiple myeloma. Proc Natl Acad Sci U S A. 1996;93:13931–13936. doi: 10.1073/pnas.93.24.13931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weinstein IB. Cancer. Addiction to oncogenes--the Achilles heal of cancer. Science. 2002;297:63–64. doi: 10.1126/science.1073096. [DOI] [PubMed] [Google Scholar]
- 9.Solimini NL, Luo J, Elledge SJ. Non-oncogene addiction and the stress phenotype of cancer cells. Cell. 2007;130:986–988. doi: 10.1016/j.cell.2007.09.007. [DOI] [PubMed] [Google Scholar]
- 10.Garraway LA, Sellers WR. Lineage dependency and lineage-survival oncogenes in human cancer. Nature reviews Cancer. 2006;6:593–602. doi: 10.1038/nrc1947. [DOI] [PubMed] [Google Scholar]
- 11.Staudt LM. Oncogenic activation of NF-kappaB. Cold Spring Harb Perspect Biol. 2010;2:a000109. doi: 10.1101/cshperspect.a000109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vallabhapurapu S, Karin M. Regulation and function of NF-kappaB transcription factors in the immune system. Annual review of immunology. 2009;27:693–733. doi: 10.1146/annurev.immunol.021908.132641. [DOI] [PubMed] [Google Scholar]
- 13.Davis RE, Brown KD, Siebenlist U, Staudt LM. Constitutive nuclear factor kappa B activity is required for survival of activated B Cell-like diffuse large B cell lymphoma cells. J Exp Med. 2001;194:1861–1874. doi: 10.1084/jem.194.12.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ngo VN, et al. A loss-of-function RNA interference screen for molecular targets in cancer. Nature. 2006;441:106–110. doi: 10.1038/nature04687. [DOI] [PubMed] [Google Scholar]
- 15.Thome M, Charton JE, Pelzer C, Hailfinger S. Antigen receptor signaling to NF-kappaB via CARMA1, BCL10, and MALT1. Cold Spring Harb Perspect Biol. 2010;2:a003004. doi: 10.1101/cshperspect.a003004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qiao Q, et al. Structural architecture of the CARMA1/Bcl10/MALT1 signalosome: nucleation-induced filamentous assembly. Molecular cell. 2013;51:766–779. doi: 10.1016/j.molcel.2013.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lenz G, et al. Oncogenic CARD11 mutations in human diffuse large B cell lymphoma. Science. 2008;319:1676–1679. doi: 10.1126/science.1153629. [DOI] [PubMed] [Google Scholar]
- 18.Davis RE, et al. Chronic active B-cell-receptor signalling in diffuse large B-cell lymphoma. Nature. 2010;463:88–92. doi: 10.1038/nature08638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ngo VN, et al. Oncogenically active MYD88 mutations in human lymphoma. Nature. 2011;470:115–119. doi: 10.1038/nature09671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Puente XS, et al. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature. 2011 doi: 10.1038/nature10113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Treon SP, et al. MYD88 L265P somatic mutation in Waldenstrom’s macroglobulinemia. N Engl J Med. 2012;367:826–833. doi: 10.1056/NEJMoa1200710. [DOI] [PubMed] [Google Scholar]
- 22.Bhoj VG, Chen ZJ. Ubiquitylation in innate and adaptive immunity. Nature. 2009;458:430–437. doi: 10.1038/nature07959. [DOI] [PubMed] [Google Scholar]
- 23.Hershko A, Ciechanover A. The ubiquitin system. Annual review of biochemistry. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 24.Kirisako T, et al. A ubiquitin ligase complex assembles linear polyubiquitin chains. The EMBO journal. 2006;25:4877–4887. doi: 10.1038/sj.emboj.7601360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fraile JM, Quesada V, Rodriguez D, Freije JM, Lopez-Otin C. Deubiquitinases in cancer: new functions and therapeutic options. Oncogene. 2012;31:2373–2388. doi: 10.1038/onc.2011.443. [DOI] [PubMed] [Google Scholar]
- 26.Ikeda F, Crosetto N, Dikic I. What determines the specificity and outcomes of ubiquitin signaling? Cell. 2010;143:677–681. doi: 10.1016/j.cell.2010.10.026. [DOI] [PubMed] [Google Scholar]
- 27.Thrower JS, Hoffman L, Rechsteiner M, Pickart CM. Recognition of the polyubiquitin proteolytic signal. The EMBO journal. 2000;19:94–102. doi: 10.1093/emboj/19.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jin L, Williamson A, Banerjee S, Philipp I, Rape M. Mechanism of ubiquitin-chain formation by the human anaphase-promoting complex. Cell. 2008;133:653–665. doi: 10.1016/j.cell.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun L, Chen ZJ. The novel functions of ubiquitination in signaling. Curr Opin Cell Biol. 2004;16:119–126. doi: 10.1016/j.ceb.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 30.Wu ZH, Shi Y. When ubiquitin meets NF-kappaB: a trove for anti-cancer drug development. Current pharmaceutical design. 2013;19:3263–3275. doi: 10.2174/1381612811319180010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gerlach B, et al. Linear ubiquitination prevents inflammation and regulates immune signalling. Nature. 2011;471:591–596. doi: 10.1038/nature09816. [DOI] [PubMed] [Google Scholar]
- 32.Ikeda F, et al. SHARPIN forms a linear ubiquitin ligase complex regulating NF-kappaB activity and apoptosis. Nature. 2011;471:637–641. doi: 10.1038/nature09814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tokunaga F, et al. SHARPIN is a component of the NF-kappaB-activating linear ubiquitin chain assembly complex. Nature. 2011;471:633–636. doi: 10.1038/nature09815. [DOI] [PubMed] [Google Scholar]
- 34.Compagno M, et al. Mutations of multiple genes cause deregulation of NF-kappaB in diffuse large B-cell lymphoma. Nature. 2009;459:717–721. doi: 10.1038/nature07968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Honma K, et al. TNFAIP3/A20 functions as a novel tumor suppressor gene in several subtypes of non-Hodgkin lymphomas. Blood. 2009;114:2467–2475. doi: 10.1182/blood-2008-12-194852. [DOI] [PubMed] [Google Scholar]
- 36.Tavares RM, et al. The ubiquitin modifying enzyme A20 restricts B cell survival and prevents autoimmunity. Immunity. 2010;33:181–191. doi: 10.1016/j.immuni.2010.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Srinivasula SM, Ashwell JD. A20: more than one way to skin a cat. Mol Cell. 2011;44:511–512. doi: 10.1016/j.molcel.2011.11.004. [DOI] [PubMed] [Google Scholar]
- 38.Zapata JM. TNF-receptor-associated factors as targets for drug development. Expert opinion on therapeutic targets. 2003;7:411–425. doi: 10.1517/14728222.7.3.411. [DOI] [PubMed] [Google Scholar]
- 39.Sun L, Deng L, Ea CK, Xia ZP, Chen ZJ. The TRAF6 ubiquitin ligase and TAK1 kinase mediate IKK activation by BCL10 and MALT1 in T lymphocytes. Molecular cell. 2004;14:289–301. doi: 10.1016/s1097-2765(04)00236-9. [DOI] [PubMed] [Google Scholar]
- 40.Oeckinghaus A, et al. Malt1 ubiquitination triggers NF-kappaB signaling upon T-cell activation. The EMBO journal. 2007;26:4634–4645. doi: 10.1038/sj.emboj.7601897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu CJ, Ashwell JD. NEMO recognition of ubiquitinated Bcl10 is required for T cell receptor-mediated NF-kappaB activation. Proc Natl Acad Sci U S A. 2008;105:3023–3028. doi: 10.1073/pnas.0712313105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ferch U, et al. Inhibition of MALT1 protease activity is selectively toxic for activated B cell-like diffuse large B cell lymphoma cells. J Exp Med. 2009;206:2313–2320. doi: 10.1084/jem.20091167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hailfinger S, et al. Essential role of MALT1 protease activity in activated B cell-like diffuse large B-cell lymphoma. Proc Natl Acad Sci U S A. 2009;106:19946–19951. doi: 10.1073/pnas.0907511106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Coornaert B, et al. T cell antigen receptor stimulation induces MALT1 paracaspase-mediated cleavage of the NF-kappaB inhibitor A20. Nature immunology. 2008;9:263–271. doi: 10.1038/ni1561. [DOI] [PubMed] [Google Scholar]
- 45.Rebeaud F, et al. The proteolytic activity of the paracaspase MALT1 is key in T cell activation. Nature immunology. 2008;9:272–281. doi: 10.1038/ni1568. [DOI] [PubMed] [Google Scholar]
- 46.Hailfinger S, et al. Malt1-dependent RelB cleavage promotes canonical NF-{kappa}B activation in lymphocytes and lymphoma cell lines. Proc Natl Acad Sci U S A. 2011;108:14596–14601. doi: 10.1073/pnas.1105020108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pelzer C, Cabalzar K, Wolf A, Gonzalez M, Lenz G, Thome M. The protease activity of the paracaspase MALT1 is controlled by monoubiquitination. Nat Immunol. 2013;14:337–345. doi: 10.1038/ni.2540. [DOI] [PubMed] [Google Scholar]
- 48.Cabalzar K, et al. Monoubiquitination and activity of the paracaspase MALT1 requires glutamate 549 in the dimerization interface. PloS one. 2013;8:e72051. doi: 10.1371/journal.pone.0072051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen J, Chen ZJ. Regulation of NF-kappaB by ubiquitination. Current opinion in immunology. 2013;25:4–12. doi: 10.1016/j.coi.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Alam SL, et al. Ubiquitin interactions of NZF zinc fingers. The EMBO journal. 2004;23:1411–1421. doi: 10.1038/sj.emboj.7600114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lamothe B, Besse A, Campos AD, Webster WK, Wu H, Darnay BG. Site-specific Lys-63-linked tumor necrosis factor receptor-associated factor 6 auto-ubiquitination is a critical determinant of I kappa B kinase activation. The Journal of biological chemistry. 2007;282:4102–4112. doi: 10.1074/jbc.M609503200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Besse A, et al. TAK1-dependent signaling requires functional interaction with TAB2/TAB3. The Journal of biological chemistry. 2007;282:3918–3928. doi: 10.1074/jbc.M608867200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ordureau A, et al. The IRAK-catalysed activation of the E3 ligase function of Pellino isoforms induces the Lys63-linked polyubiquitination of IRAK1. Biochem J. 2008;409:43–52. doi: 10.1042/BJ20071365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Newton K, et al. Ubiquitin chain editing revealed by polyubiquitin linkage-specific antibodies. Cell. 2008;134:668–678. doi: 10.1016/j.cell.2008.07.039. [DOI] [PubMed] [Google Scholar]
- 55.Conze DB, Wu CJ, Thomas JA, Landstrom A, Ashwell JD. Lys63-linked polyubiquitination of IRAK-1 is required for interleukin-1 receptor- and toll-like receptor-mediated NF-kappaB activation. Mol Cell Biol. 2008;28:3538–3547. doi: 10.1128/MCB.02098-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tokunaga F, et al. Involvement of linear polyubiquitylation of NEMO in NF-kappaB activation. Nature cell biology. 2009;11:123–132. doi: 10.1038/ncb1821. [DOI] [PubMed] [Google Scholar]
- 57.Hostager BS, et al. HOIL-1L interacting protein (HOIP) as an NF-kappaB regulating component of the CD40 signaling complex. PloS one. 2010;5:e11380. doi: 10.1371/journal.pone.0011380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Haas TL, et al. Recruitment of the linear ubiquitin chain assembly complex stabilizes the TNF-R1 signaling complex and is required for TNF-mediated gene induction. Molecular cell. 2009;36:831–844. doi: 10.1016/j.molcel.2009.10.013. [DOI] [PubMed] [Google Scholar]
- 59.Fujita H, et al. Mechanism underlying IkappaB kinase activation mediated by the linear ubiquitin chain assembly complex. Mol Cell Biol. 2014;34:1322–1335. doi: 10.1128/MCB.01538-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rahighi S, et al. Specific recognition of linear ubiquitin chains by NEMO is important for NF-kappaB activation. Cell. 2009;136:1098–1109. doi: 10.1016/j.cell.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 61.Yang Y, et al. Essential role of the linear ubiquitin chain assembly complex in lymphoma revealed by rare germline polymorphisms. Cancer discovery. 2014;4:480–493. doi: 10.1158/2159-8290.CD-13-0915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dubois SM, et al. A catalytic-independent role for the LUBAC in NF-kappaB activation upon antigen receptor engagement and in lymphoma cells. Blood. 2014;123:2199–2203. doi: 10.1182/blood-2013-05-504019. [DOI] [PubMed] [Google Scholar]
- 63.Sasaki Y, et al. Defective immune responses in mice lacking LUBAC-mediated linear ubiquitination in B cells. The EMBO journal. 2013;32:2463–2476. doi: 10.1038/emboj.2013.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yagi H, et al. A non-canonical UBA-UBL interaction forms the linear-ubiquitin-chain assembly complex. EMBO reports. 2012;13:462–468. doi: 10.1038/embor.2012.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Boisson B, et al. Immunodeficiency, autoinflammation and amylopectinosis in humans with inherited HOIL-1 and LUBAC deficiency. Nat Immunol. 2012;13:1178–1186. doi: 10.1038/ni.2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nijman SM, et al. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123:773–786. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 67.Wertz IE, et al. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-kappaB signalling. Nature. 2004;430:694–699. doi: 10.1038/nature02794. [DOI] [PubMed] [Google Scholar]
- 68.Skaug B, Chen J, Du F, He J, Ma A, Chen ZJ. Direct, noncatalytic mechanism of IKK inhibition by A20. Mol Cell. 2011;44:559–571. doi: 10.1016/j.molcel.2011.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bosanac I, et al. Ubiquitin binding to A20 ZnF4 is required for modulation of NF-kappaB signaling. Mol Cell. 2010;40:548–557. doi: 10.1016/j.molcel.2010.10.009. [DOI] [PubMed] [Google Scholar]
- 70.Schmitz R, et al. TNFAIP3 (A20) is a tumor suppressor gene in Hodgkin lymphoma and primary mediastinal B cell lymphoma. J Exp Med. 2009;206:981–989. doi: 10.1084/jem.20090528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kato M, et al. Frequent inactivation of A20 in B-cell lymphomas. Nature. 2009;459:712–716. doi: 10.1038/nature07969. [DOI] [PubMed] [Google Scholar]
- 72.Novak U, et al. The NF-{kappa}B negative regulator TNFAIP3 (A20) is inactivated by somatic mutations and genomic deletions in marginal zone B-cell lymphomas. Blood. 2009 doi: 10.1182/blood-2008-08-174110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lin SC, et al. Molecular basis for the unique deubiquitinating activity of the NF-kappaB inhibitor A20. J Mol Biol. 2008;376:526–540. doi: 10.1016/j.jmb.2007.11.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hymowitz SG, Wertz IE. A20: from ubiquitin editing to tumour suppression. Nat Rev Cancer. 2010;10:332–341. doi: 10.1038/nrc2775. [DOI] [PubMed] [Google Scholar]
- 75.Tokunaga F, et al. Specific recognition of linear polyubiquitin by A20 zinc finger 7 is involved in NF-kappaB regulation. The EMBO journal. 2012;31:3856–3870. doi: 10.1038/emboj.2012.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Verhelst K, et al. A20 inhibits LUBAC-mediated NF-kappaB activation by binding linear polyubiquitin chains via its zinc finger 7. The EMBO journal. 2012;31:3845–3855. doi: 10.1038/emboj.2012.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sun SC. CYLD: a tumor suppressor deubiquitinase regulating NF-kappaB activation and diverse biological processes. Cell death and differentiation. 2010;17:25–34. doi: 10.1038/cdd.2009.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Brummelkamp TR, Nijman SM, Dirac AM, Bernards R. Loss of the cylindromatosis tumour suppressor inhibits apoptosis by activating NF-kappaB. Nature. 2003;424:797–801. doi: 10.1038/nature01811. [DOI] [PubMed] [Google Scholar]
- 79.Kovalenko A, Chable-Bessia C, Cantarella G, Israel A, Wallach D, Courtois G. The tumour suppressor CYLD negatively regulates NF-kappaB signalling by deubiquitination. Nature. 2003;424:801–805. doi: 10.1038/nature01802. [DOI] [PubMed] [Google Scholar]
- 80.Trompouki E, Hatzivassiliou E, Tsichritzis T, Farmer H, Ashworth A, Mosialos G. CYLD is a deubiquitinating enzyme that negatively regulates NF-kappaB activation by TNFR family members. Nature. 2003;424:793–796. doi: 10.1038/nature01803. [DOI] [PubMed] [Google Scholar]
- 81.Staal J, et al. T-cell receptor-induced JNK activation requires proteolytic inactivation of CYLD by MALT1. The EMBO journal. 2011;30:1742–1752. doi: 10.1038/emboj.2011.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Annunziata CM, et al. Frequent engagement of the classical and alternative NF-kappaB pathways by diverse genetic abnormalities in multiple myeloma. Cancer cell. 2007;12:115–130. doi: 10.1016/j.ccr.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Keats JJ, et al. Promiscuous mutations activate the noncanonical NF-kappaB pathway in multiple myeloma. Cancer cell. 2007;12:131–144. doi: 10.1016/j.ccr.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ye Y, Rape M. Building ubiquitin chains: E2 enzymes at work. Nature reviews. Molecular cell biology. 2009;10:755–764. doi: 10.1038/nrm2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wertz IE, Dixit VM. Signaling to NF-kappaB: regulation by ubiquitination. Cold Spring Harb Perspect Biol. 2010;2:a003350. doi: 10.1101/cshperspect.a003350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 87.Torres-Ramos CA, Prakash S, Prakash L. Requirement of RAD5 and MMS2 for postreplication repair of UV-damaged DNA in Saccharomyces cerevisiae. Molecular and cellular biology. 2002;22:2419–2426. doi: 10.1128/MCB.22.7.2419-2426.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Xiao W, Chow BL, Broomfield S, Hanna M. The Saccharomyces cerevisiae RAD6 group is composed of an error-prone and two error-free postreplication repair pathways. Genetics. 2000;155:1633–1641. doi: 10.1093/genetics/155.4.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Pulvino M, et al. Inhibition of proliferation and survival of diffuse large B-cell lymphoma cells by a small-molecule inhibitor of the ubiquitin-conjugating enzyme Ubc13-Uev1A. Blood. 2012;120:1668–1677. doi: 10.1182/blood-2012-02-406074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Jin J, Cardozo T, Lovering RC, Elledge SJ, Pagano M, Harper JW. Systematic analysis and nomenclature of mammalian F-box proteins. Genes & development. 2004;18:2573–2580. doi: 10.1101/gad.1255304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Nakayama KI, Nakayama K. Regulation of the cell cycle by SCF-type ubiquitin ligases. Seminars in cell & developmental biology. 2005;16:323–333. doi: 10.1016/j.semcdb.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 92.Brown K, Gerstberger S, Carlson L, Franzoso G, Siebenlist U. Control of I kappa B-alpha proteolysis by site-specific, signal-induced phosphorylation. Science. 1995;267:1485–1488. doi: 10.1126/science.7878466. [DOI] [PubMed] [Google Scholar]
- 93.Yaron A, et al. Identification of the receptor component of the IkappaBalpha-ubiquitin ligase. Nature. 1998;396:590–594. doi: 10.1038/25159. [DOI] [PubMed] [Google Scholar]
- 94.Liu J, et al. Oncogenic BRAF regulates beta-Trcp expression and NF-kappaB activity in human melanoma cells. Oncogene. 2007;26:1954–1958. doi: 10.1038/sj.onc.1209994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Dhawan P, Richmond A. A novel NF-kappa B-inducing kinase-MAPK signaling pathway up-regulates NF-kappa B activity in melanoma cells. The Journal of biological chemistry. 2002;277:7920–7928. doi: 10.1074/jbc.M112210200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Milhollen MA, et al. MLN4924, a NEDD8-activating enzyme inhibitor, is active in diffuse large B-cell lymphoma models: rationale for treatment of NF-{kappa}B-dependent lymphoma. Blood. 2010;116:1515–1523. doi: 10.1182/blood-2010-03-272567. [DOI] [PubMed] [Google Scholar]
- 97.Wang Z, Liu P, Inuzuka H, Wei W. Roles of F-box proteins in cancer. Nature reviews Cancer. 2014;14:233–247. doi: 10.1038/nrc3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chiorazzi M, et al. Related F-box proteins control cell death in Caenorhabditis elegans and human lymphoma. Proc Natl Acad Sci U S A. 2013;110:3943–3948. doi: 10.1073/pnas.1217271110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Duan S, et al. FBXO11 targets BCL6 for degradation and is inactivated in diffuse large B-cell lymphomas. Nature. 2012;481:90–93. doi: 10.1038/nature10688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Basso K, Dalla-Favera R. Roles of BCL6 in normal and transformed germinal center B cells. Immunol Rev. 2012;247:172–183. doi: 10.1111/j.1600-065X.2012.01112.x. [DOI] [PubMed] [Google Scholar]
- 101.Niu H, Ye BH, Dalla-Favera R. Antigen receptor signaling induces MAP kinase-mediated phosphorylation and degradation of the BCL-6 transcription factor. Genes & development. 1998;12:1953–1961. doi: 10.1101/gad.12.13.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Phan RT, Saito M, Kitagawa Y, Means AR, Dalla-Favera R. Genotoxic stress regulates expression of the proto-oncogene Bcl6 in germinal center B cells. Nature immunology. 2007;8:1132–1139. doi: 10.1038/ni1508. [DOI] [PubMed] [Google Scholar]
- 103.Dimmeler S, Breitschopf K, Haendeler J, Zeiher AM. Dephosphorylation targets Bcl-2 for ubiquitin-dependent degradation: a link between the apoptosome and the proteasome pathway. J Exp Med. 1999;189:1815–1822. doi: 10.1084/jem.189.11.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Breitschopf K, Haendeler J, Malchow P, Zeiher AM, Dimmeler S. Posttranslational modification of Bcl-2 facilitates its proteasome-dependent degradation: molecular characterization of the involved signaling pathway. Molecular and cellular biology. 2000;20:1886–1896. doi: 10.1128/mcb.20.5.1886-1896.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lin SS, et al. PP2A regulates BCL-2 phosphorylation and proteasome-mediated degradation at the endoplasmic reticulum. The Journal of biological chemistry. 2006;281:23003–23012. doi: 10.1074/jbc.M602648200. [DOI] [PubMed] [Google Scholar]
- 106.Basu A, DuBois G, Haldar S. Posttranslational modifications of Bcl2 family members--a potential therapeutic target for human malignancy. Frontiers in bioscience : a journal and virtual library. 2006;11:1508–1521. doi: 10.2741/1900. [DOI] [PubMed] [Google Scholar]
- 107.Hao B, Oehlmann S, Sowa ME, Harper JW, Pavletich NP. Structure of a Fbw7-Skp1-cyclin E complex: multisite-phosphorylated substrate recognition by SCF ubiquitin ligases. Molecular cell. 2007;26:131–143. doi: 10.1016/j.molcel.2007.02.022. [DOI] [PubMed] [Google Scholar]
- 108.Orlicky S, Tang X, Willems A, Tyers M, Sicheri F. Structural basis for phosphodependent substrate selection and orientation by the SCFCdc4 ubiquitin ligase. Cell. 2003;112:243–256. doi: 10.1016/s0092-8674(03)00034-5. [DOI] [PubMed] [Google Scholar]
- 109.Welcker M, Clurman BE. FBW7 ubiquitin ligase: a tumour suppressor at the crossroads of cell division, growth and differentiation. Nature reviews Cancer. 2008;8:83–93. doi: 10.1038/nrc2290. [DOI] [PubMed] [Google Scholar]
- 110.Minella AC, Clurman BE. Mechanisms of tumor suppression by the SCF(Fbw7) Cell cycle. 2005;4:1356–1359. doi: 10.4161/cc.4.10.2058. [DOI] [PubMed] [Google Scholar]
- 111.Maser RS, et al. Chromosomally unstable mouse tumours have genomic alterations similar to diverse human cancers. Nature. 2007;447:966–971. doi: 10.1038/nature05886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Crusio KM, King B, Reavie LB, Aifantis I. The ubiquitous nature of cancer: the role of the SCF(Fbw7) complex in development and transformation. Oncogene. 2010;29:4865–4873. doi: 10.1038/onc.2010.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Weng AP, et al. c-Myc is an important direct target of Notch1 in T-cell acute lymphoblastic leukemia/lymphoma. Genes & development. 2006;20:2096–2109. doi: 10.1101/gad.1450406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.O’Neil J, et al. FBW7 mutations in leukemic cells mediate NOTCH pathway activation and resistance to gamma-secretase inhibitors. J Exp Med. 2007;204:1813–1824. doi: 10.1084/jem.20070876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Weng AP, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269–271. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- 116.King B, et al. The ubiquitin ligase FBXW7 modulates leukemia-initiating cell activity by regulating MYC stability. Cell. 2013;153:1552–1566. doi: 10.1016/j.cell.2013.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Thompson BJ, et al. The SCFFBW7 ubiquitin ligase complex as a tumor suppressor in T cell leukemia. J Exp Med. 2007;204:1825–1835. doi: 10.1084/jem.20070872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Busino L, et al. Fbxw7alpha- and GSK3-mediated degradation of p100 is a pro-survival mechanism in multiple myeloma. Nature cell biology. 2012;14:375–385. doi: 10.1038/ncb2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Basak S, et al. A fourth IkappaB protein within the NF-kappaB signaling module. Cell. 2007;128:369–381. doi: 10.1016/j.cell.2006.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Angers S, Li T, Yi X, MacCoss MJ, Moon RT, Zheng N. Molecular architecture and assembly of the DDB1-CUL4A ubiquitin ligase machinery. Nature. 2006;443:590–593. doi: 10.1038/nature05175. [DOI] [PubMed] [Google Scholar]
- 121.He YJ, McCall CM, Hu J, Zeng Y, Xiong Y. DDB1 functions as a linker to recruit receptor WD40 proteins to CUL4-ROC1 ubiquitin ligases. Genes & development. 2006;20:2949–2954. doi: 10.1101/gad.1483206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Higa LA, Wu M, Ye T, Kobayashi R, Sun H, Zhang H. CUL4-DDB1 ubiquitin ligase interacts with multiple WD40-repeat proteins and regulates histone methylation. Nature cell biology. 2006;8:1277–1283. doi: 10.1038/ncb1490. [DOI] [PubMed] [Google Scholar]
- 123.Jin J, Arias EE, Chen J, Harper JW, Walter JC. A family of diverse Cul4-Ddb1-interacting proteins includes Cdt2, which is required for S phase destruction of the replication factor Cdt1. Molecular cell. 2006;23:709–721. doi: 10.1016/j.molcel.2006.08.010. [DOI] [PubMed] [Google Scholar]
- 124.Ito T, et al. Identification of a primary target of thalidomide teratogenicity. Science. 2010;327:1345–1350. doi: 10.1126/science.1177319. [DOI] [PubMed] [Google Scholar]
- 125.Higgins JJ, Rosen DR, Loveless JM, Clyman JC, Grau MJ. A gene for nonsyndromic mental retardation maps to chromosome 3p25-pter. Neurology. 2000;55:335–340. doi: 10.1212/wnl.55.3.335. [DOI] [PubMed] [Google Scholar]
- 126.Dijkhuizen T, et al. FISH and array-CGH analysis of a complex chromosome 3 aberration suggests that loss of CNTN4 and CRBN contributes to mental retardation in 3pter deletions. American journal of medical genetics Part A. 2006;140:2482–2487. doi: 10.1002/ajmg.a.31487. [DOI] [PubMed] [Google Scholar]
- 127.Higgins JJ, Pucilowska J, Lombardi RQ, Rooney JP. A mutation in a novel ATP-dependent Lon protease gene in a kindred with mild mental retardation. Neurology. 2004;63:1927–1931. doi: 10.1212/01.wnl.0000146196.01316.a2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Jo S, Lee KH, Song S, Jung YK, Park CS. Identification and functional characterization of cereblon as a binding protein for large-conductance calcium-activated potassium channel in rat brain. Journal of neurochemistry. 2005;94:1212–1224. doi: 10.1111/j.1471-4159.2005.03344.x. [DOI] [PubMed] [Google Scholar]
- 129.Higgins JJ, et al. Temporal and spatial mouse brain expression of cereblon, an ionic channel regulator involved in human intelligence. Journal of neurogenetics. 2010;24:18–26. doi: 10.3109/01677060903567849. [DOI] [PubMed] [Google Scholar]
- 130.Hohberger B, Enz R. Cereblon is expressed in the retina and binds to voltage-gated chloride channels. FEBS letters. 2009;583:633–637. doi: 10.1016/j.febslet.2009.01.018. [DOI] [PubMed] [Google Scholar]
- 131.Lee KM, Jo S, Kim H, Lee J, Park CS. Functional modulation of AMP-activated protein kinase by cereblon. Biochimica et biophysica acta. 2011;1813:448–455. doi: 10.1016/j.bbamcr.2011.01.005. [DOI] [PubMed] [Google Scholar]
- 132.Zhu YX, et al. Cereblon expression is required for the antimyeloma activity of lenalidomide and pomalidomide. Blood. 2011;118:4771–4779. doi: 10.1182/blood-2011-05-356063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Yang Y, et al. Exploiting synthetic lethality for the therapy of ABC diffuse large B cell lymphoma. Cancer cell. 2012;21:723–737. doi: 10.1016/j.ccr.2012.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Lopez-Girona A, et al. Cereblon is a direct protein target for immunomodulatory and antiproliferative activities of lenalidomide and pomalidomide. Leukemia. 2012;26:2326–2335. doi: 10.1038/leu.2012.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Shaffer AL, et al. IRF4 addiction in multiple myeloma. Nature. 2008;454:226–231. doi: 10.1038/nature07064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Hernandez-Ilizaliturri FJ, et al. Higher response to lenalidomide in relapsed/refractory diffuse large b-cell lymphoma in nongerminal center b-cell-like than in germinal center b-cell-like phenotype. Cancer. 2011 doi: 10.1002/cncr.26135. [DOI] [PubMed] [Google Scholar]
- 137.Gandhi AK, et al. Immunomodulatory agents lenalidomide and pomalidomide co-stimulate T cells by inducing degradation of T cell repressors Ikaros and Aiolos via modulation of the E3 ubiquitin ligase complex CRL4(CRBN.) British journal of haematology. 2014;164:811–821. doi: 10.1111/bjh.12708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kronke J, et al. Lenalidomide causes selective degradation of IKZF1 and IKZF3 in multiple myeloma cells. Science. 2014;343:301–305. doi: 10.1126/science.1244851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Lu G, et al. The myeloma drug lenalidomide promotes the cereblon-dependent destruction of Ikaros proteins. Science. 2014;343:305–309. doi: 10.1126/science.1244917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Fischer ES, et al. Structure of the DDB1-CRBN E3 ubiquitin ligase in complex with thalidomide. Nature. 2014;512:49–53. doi: 10.1038/nature13527. [DOI] [PMC free article] [PubMed] [Google Scholar]