Figure 2. Phylogeny of hantaviruses hosted by Chiroptera, Soricomorpha or Rodents.
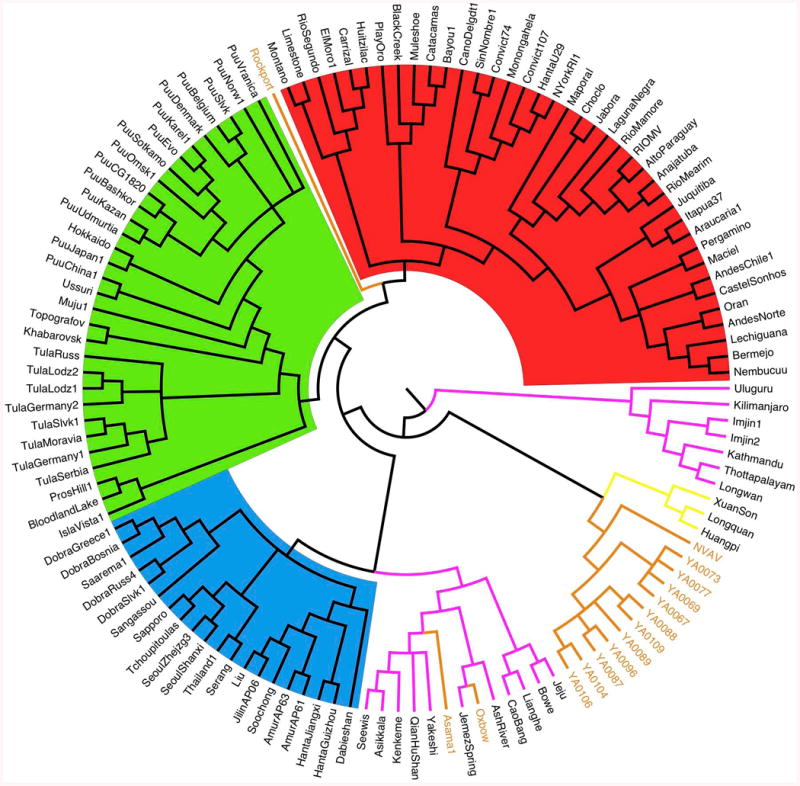
Cladogram resulting of Bayesian (MCMC) analysis (GTR+I+G model), based on the entire coding region of the S gene (nucleotides 43–1341). Hantaviruses hosted by different host groups are highlighted in: red (Sigmodontinae-Neotominae), green (Arvicolinae), blue (Murinae), pink (Soricidae), light brown (Talpidae) and yellow (Chiroptera). The analysis used 1) an extent data set of the S gene coding region available on GenBank, including most of the hantavirus strains detected either in rodents or non-rodent mammals (Soricomorpha and Chiroptera), and 2) 11 complete S-segment sequences from moles captured either in Ozoir (4) or Bauvais (7). GenBank accession numbers: FJ539168, NVAV from Hungary; KF010573–KF010576, NVAV strains from Ozoir-la-Ferrière; KF010565–KF010571, NVAV strains from Beauvais. For Chiroptera and Soricomorpha hosts, see Table 1; for rodent hosts, see Herbreteau et al., 2007, Table 16.1.
