Figure 3.
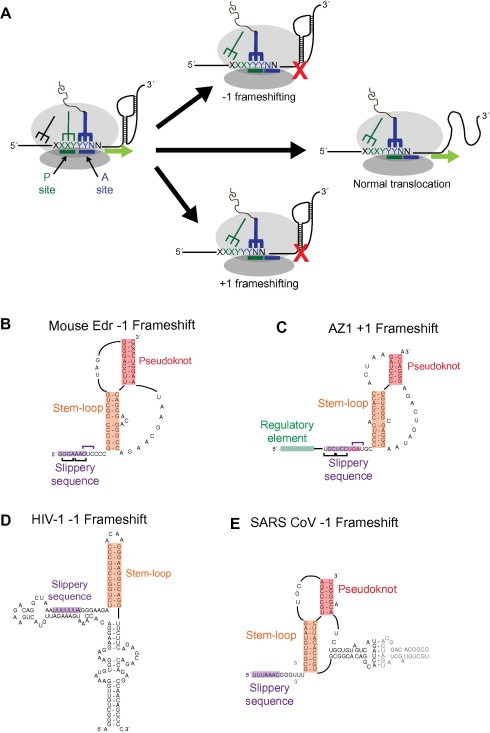
Structural diversity within programmed ribosomal frameshifting elements. (A) Schematic diagram of −1 and +1 programmed frameshifting events. A translating ribosome encounters a slippery sequence upstream of a stable RNA structure. The post‐translocation position of the peptide‐linked tRNA (blue structure) is shown for normal translocation (middle) and −1 and +1 frameshifting events. Proposed secondary structures for (B) the −1 frameshift element for the mouse Edr gene [73], (C) the +1 frameshifting region of the human AZ1 gene [76, 79] (D) the gag‐pol frameshift in HIV‐1 [82], and (E) the replicase‐transcriptase frameshift of SARS‐CoV [84]. In each example, the “slippery sequence” (purple box) is located upstream of a stable structure (orange and red boxes).
