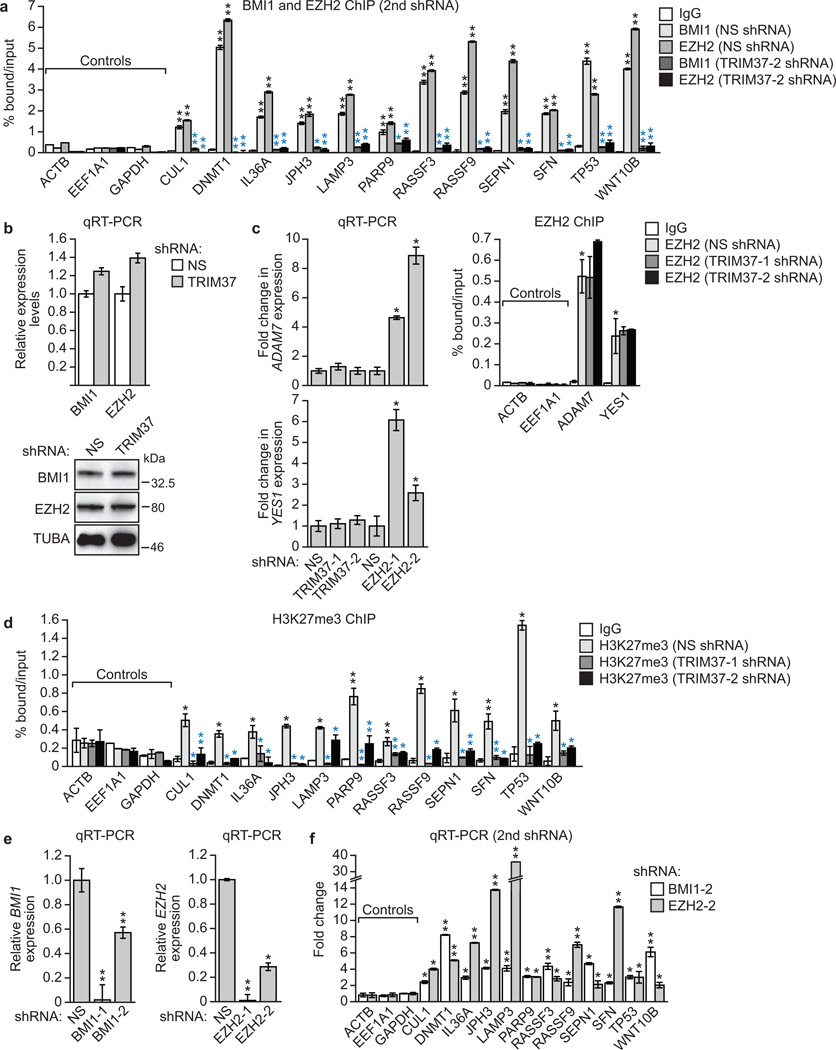
Extended Data Fig. 5. Confirmation of the results of Fig. 3f, h using second, unrelated shRNAs.
a, ChIP monitoring binding of BMI1 and EZH2 to the promoters of TRIM37 target genes in MCF7 cells expressing a NS shRNA or a second TRIM37 shRNA unrelated to that used in Fig. 3f. The IgG control and BMI1 and EZH2 signal in cells expressing a NS shRNA are the same as those shown in Fig. 3f. Black asterisks indicate significance of BMI1 or EZH2 enrichment compared to the IgG control (from cells expressing a NS shRNA); blue asterisks indicate significant differences in BMI1 or EZH2 enrichment in cells expressing a TRIM37 shRNA relative to NS shRNA. Error bars indicate SD; n=3 technical replicates of a representative experiment (out of three experiments). b, qRT-PCR (top) and immunoblot (bottom) monitoring BMI1 and EZH2 levels in MCF7 cells expressing a NS or TRIM37 shRNA. Error bars indicate SEM; n=3 technical replicates of a representative experiment (out of three experiments). The results indicate that BMI1 and EZH2 levels are unaffected by TRIM37 knockdown in MCF7 cells. c, (Left) qRT-PCR analysis monitoring expression of two genes that are bound by EZH2 but not TRIM37, ADAM7 (top) and YES1 (bottom), following knockdown of TRIM37 or EZH2 in MCF7 cells. Error bars indicate SEM; n=3 technical replicates of a representative experiment (out of three experiments). The results indicate that knockdown of EZH2 but not TRIM37 de-represses ADAM7 and YES1 expression. (Right) ChIP monitoring EZH2 enrichment at the promoters of ADAM7 and YES1 in MCF7 cells expressing a NS or TRIM37 shRNA. Error bars indicate SD; n=3 technical replicates of a representative experiment (out of three experiments). The results show that knockdown of TRIM37 has no effect on EZH2 binding at ADAM7 and YES1 promoters. Thus, loss of EZH2 binding and de-repression following TRIM37 knockdown is not general to PRC2-bound genes but rather is selective for TRIM37 target genes. d, ChIP monitoring H3K27me3 enrichment at promoters of TRIM37 target genes in MCF7 cells expressing a NS or TRIM37 shRNA. Error bars indicate SD; n=3 technical replicates of a representative experiment (out of three experiments). e, qRT-PCR analysis monitoring BMI1 (left) and EZH2 (right) knockdown efficiency in MCF7 cells following shRNA-mediated knockdown using two unrelated shRNAs against each gene. Expression of each gene was normalized to that obtained with a NS shRNA, which was set to 1. Error bars indicate SEM; n=3 technical replicates of a representative experiment (out of three experiments). f, qRT-PCR monitoring TRIM37 target gene expression in MCF7 cells following knockdown of BMI1 or EZH2 using a second shRNA unrelated to that used in Fig. 3h. Error bars indicate SEM; n=3 technical replicates of a representative experiment (out of three experiments). *P<0.05; **P<0.01.