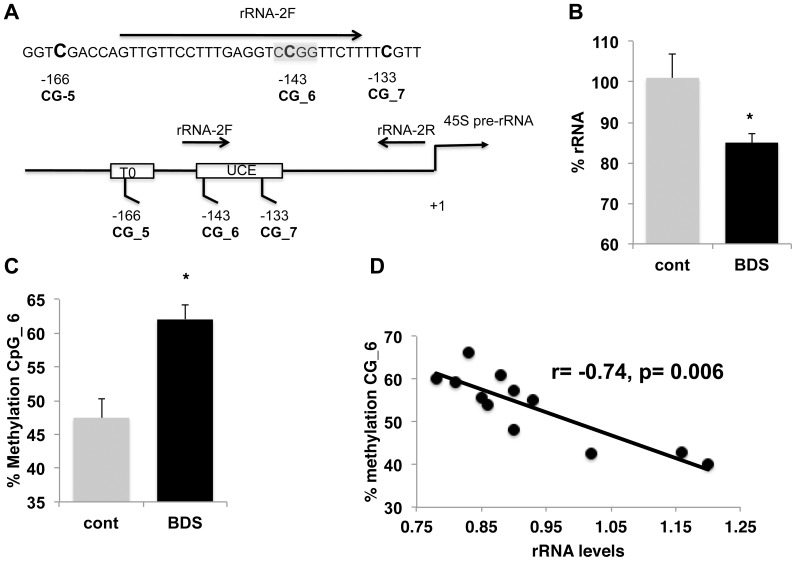
Figure 4. PND14 pups exposed to BDS show increased methylation rate at CG_6 in a manner that is highly correlated with a decrease in rRNA levels.
A (top) Nucleotide sequence of primer rRNA-2F (arrow) is shown with HpaII restriction site at CG_6 is highlighted in grey, (bottom), schematic diagram of Q-PCR primers used to assess DNA methylation at CG_6. Note that no product will be generated if CG_6 is not methylated and the DNA is digested with HpaII. Exposure to BDS decreases rRNA levels (B) while increasing DNA methylation at CG_6 in 14-day old pups (C). D. Levels of methylation at CG_6 are inversely correlated with rRNA levels in the developing hippocampus. Unpaired Student t tests (B, C), Pearson correlation (D). Error bars represent mean ± SEM, *p<0.05.