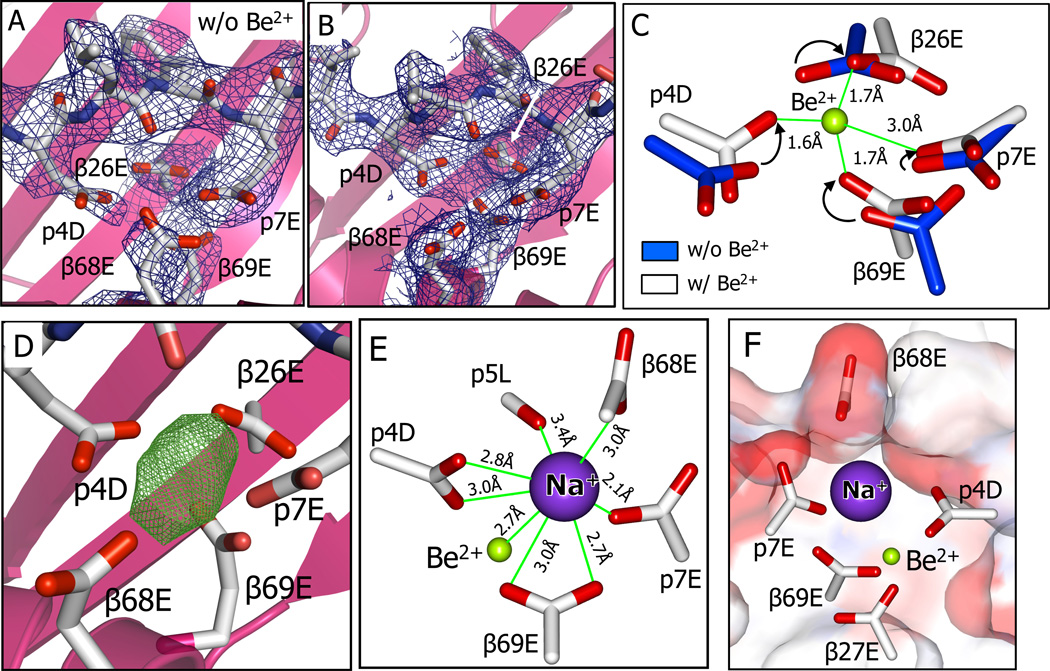
Figure 3. Be2+ and an accompanying Na+ cation captured in the acidic pocket are shielded from the solvent.
(A and B) View from above looking into the acidic pocket of DP2-M2-Be2+ with the side chains of β26E, β68E, β69E, p4D, and p7E shown with 2Fo-Fc (1σ) electron density before (A) and after (B) capture of Be2.
(C) Overlay of the carboxylates of β26E, β69E, p4D and p7E before (carbon, blue) and after (carbon, white) binding of Be2+. Arrows show the Be2+ induced movement of the relevant carboxylate oxygens. Be2+ (green) is modeled within the cluster of oxygens based on small molecule structures of Be2+ coordinated by carboxylates (Alerghi et al., 1999; Schmidbaur et al., 1991; Schmidt et al., 1997) and the Be-O bond lengths are shown.
(D) Unaccounted for electron density in the Fo-Fc (6σ) map in the acidic pocket of DP2-M2-Be2+.
(E) A Na+ cation is shown modeled into Fo-Fc density of (D). Distances of the Na+ from Be2+ and from oxygens of the carboxylates of 68E, β69E, p4D, and p7E and the backbone carbonyl oxygen of p5L are shown.
(F) A view looking through the M2 peptide toward the DP2 β1 alpha helix showing the water accessible surface of the DP2-M2-Be2+ colored by the contributing atoms (oxygen-red, carbon-white). The surface contributed by the DP2 α1 alpha helix has been clipped to reveal the carboxylates of the five acidic amino acids in the pocket along with Be2+ and Na+ among which only β68E, p4D, and p7E are surface accessible.