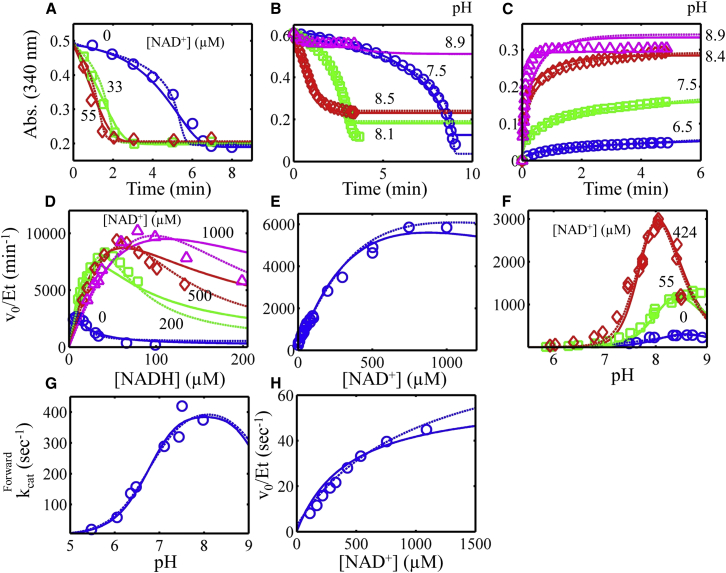
Figure 3.
Global fitting of the E. coli E3 progress curve, substrate inhibition, NAD+ activation, and pH-dependent data obtained from previous studies (3, 11, 42) (see Supporting Material). All data (symbols) were fitted to two models, where the Kd for substrates and products was either independent (solid lines) or dependent (dashed lines) on the enzyme redox state, as described in Methods. (A) Progress curves for the reverse reaction (pH 7.9) in different NAD+ concentrations, with data obtained from Fig. 1 of Wilkinson and Williams (11). (B) Progress curves for the reverse reaction, with pH varied, with data obtained from Fig. 5 of Koike et al. (3). (C) Progress curves in the forward direction with pH varied, with data obtained from Fig. 4 of Koike et al. (3). (D) Initial velocity divided by total enzyme concentration (v0/Et) in the reverse direction (pH 7.9) with varied NADH with different fixed NAD+ concentrations. Data were obtained from Fig. 6 of Wilkinson and Williams (11). (E) Activation of reverse v0/Et (pH 7.9) with increasing NAD+ concentration, with data obtained from Fig. 3 of Wilkinson and Williams (11). (F) v0/Et as a function of pH and NAD+ concentration, with data obtained from Fig. 2 of Wilkinson and Williams (11). (G) Forward kcat as a function of pH, with data obtained from Fig. 4 A of Sahlman and Williams (42). (H) v0/Et as a function of NAD+ concentration (pH 6.05), with data obtained from Fig. 2 of Sahlman and Williams (42). To see this figure in color, go online.