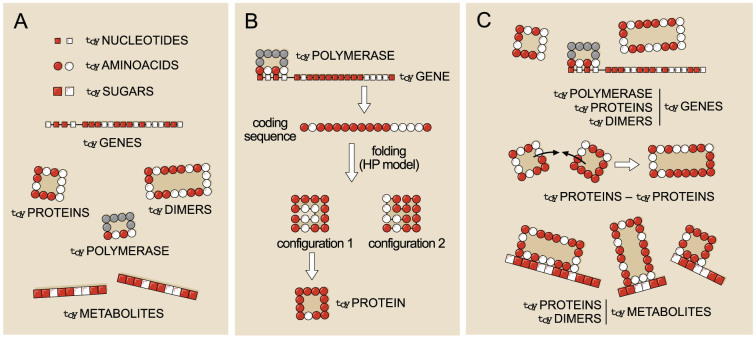
Figure 1. Building blocks and interactions defining toyLIFE.
(A): The three basic building blocks of toyLIFE are toyNucleotides, toyAminoacids, and toySugars. They can be hydrophobic (H, white) or polar (P, red), and their random polymers constitute toyGenes, toyProteins, and toyMetabolites. (B): Folding of a toyProtein. When a toyGene is expressed, its coding region is translated into a sequence of toyAminoacids, which folds on a 4 × 4 lattice following a self-avoiding walk. As a result, the toyProtein acquires a folding energy, which is the sum of the interaction energies between non-contiguous toyAminoacids of the chain (one, two or three, with energies ranging from 0 to −2). Interaction energy is pairwise additive. A toyProtein folds into the structure that minimises this folding energy. If two structures have the same minimal folding energy, the one with the minimum number of H toyAminoacids on its perimeter is chosen; if this number also coincides, the toyProtein does not fold. toyProteins are therefore characterised by two traits: their perimeter and their folding energy. (C): Possible interactions between pairs of toyLIFE elements. toyGenes interact through their promoter region with toyProteins (including the toyPolymerase and toyDimers); toyProteins can bind to form toyDimers, and interact with the toyPolymerase when bound to a promoter; both toyProteins and toyDimers can bind a toyMetabolite at arbitrary regions along its sequence.