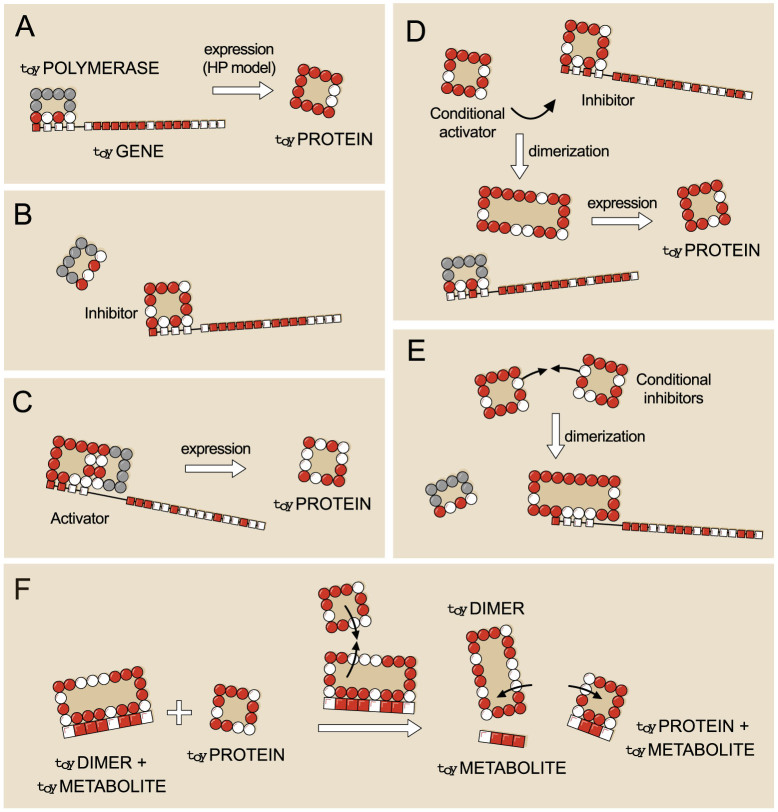
Figure 2. Regulatory and metabolic functions in toyProteins.
(A): A toyGene is expressed (translated) when the toyPolymerase binds to its promoter region. The sequence of Ps and Hs of the toyProtein will be exactly the same as that of the toyGene coding region. (B): If a toyProtein binds to the promoter region of a toyGene with a lower energy than the toyPolymerase does, it will displace the latter, and the toyGene will not be expressed. This toyProtein acts as an inhibitor. (C): The toyPolymerase does not bind to every promoter region. Thus, not all toyGenes are expressed constitutively. However, some toyProteins will be able to bind to these promoter regions. If, once bound to the promoter, they bind to the toyPolymerase with their rightmost side, the toyGene will be expressed, and these toyProteins act as activators. (D): More complex interactions —involving more elements— appear. For example, a toyProtein that forms a toyDimer with an inhibitor —preventing it from binding to the promoter— will effectively activate the expression of the toyGene. However, it does neither interact with the promoter region nor with the toyPolymerase, and its function is carried out only when the inhibitor is present. We call this kind of toyProteins conditional activators. (E): Two toyProteins can bind together to form a toyDimer that inhibits the expression of a certain toyGene. As they need each other to perform this function, we call them conditional inhibitors. As the number of genes increases, this kind of complex relationships can become very intricate. (F): Catabolism in toyLIFE. A toyDimer is bound to a toyMetabolite when a new toyProtein comes in. If the new toyProtein binds to one of the two units of the toyDimer, forming a new toyDimer energetically more stable than the old one, the two toyProteins will unbind and break the toyMetabolite up into two pieces. We say that the toyMetabolite has been catabolised.