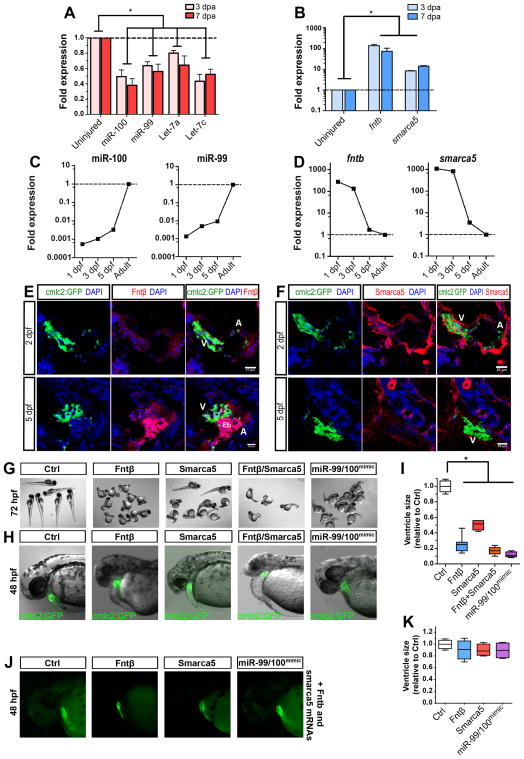
Figure 1. miR-99/100 and its direct targets contribute to zebrafishheartregeneration and development.
(A, B) Real time RT-PCR for microRNA candidates miR-99/100 and Let-7a/c (A) and downstream targets fntb and smarca5 (B) in regenerating zebrafish hearts 3 and 7 days post-amputation (dpa) (n=8). (C) Real time RT-PCR revealed that expression of miR-99/100 is very low during the first stages of development and dramatically increased at 3 days post-fertilization (dpf) in zebrafish (n=10). (D) fntb and smarca5 expression inversely correlates with miR-99/100 in developing embryos (n=10). Note that in C and D, analyzes were done in whole embryos at 1, 3, and 5dpf, and whole heart at the adult stage. (E, F) Representative pictures demonstrating that both Fntβ and Smarca5 are present at high levels in the ventricles of developing hearts at 2 and 5dpf (n=10). V: ventricle; A: atrium; Eb: erythroblasts. (G–I) Knock-down of Fntβ and/or Smarca5 in zebrafishembryos resulted in abnormally small animals (G) and reduced ventricle size in cmlc2:GFP animals (H–I). The same phenotypes were observed upon injection of miR-99/100 mimics (G–I) (n>50). Rescue experiments for the conditions tested in G were performed by co-injecting in vitro transcribed mRNAs with modified 5′UTRs with corresponding morpholinos to determine the specificity of the observed phenotypes (J) (n=30). (K) Quantification of the ventricle size for the rescue experiments. Data are represented as mean +/− s.e.m. *p<0.05. See also Figure S1 and Table S1.