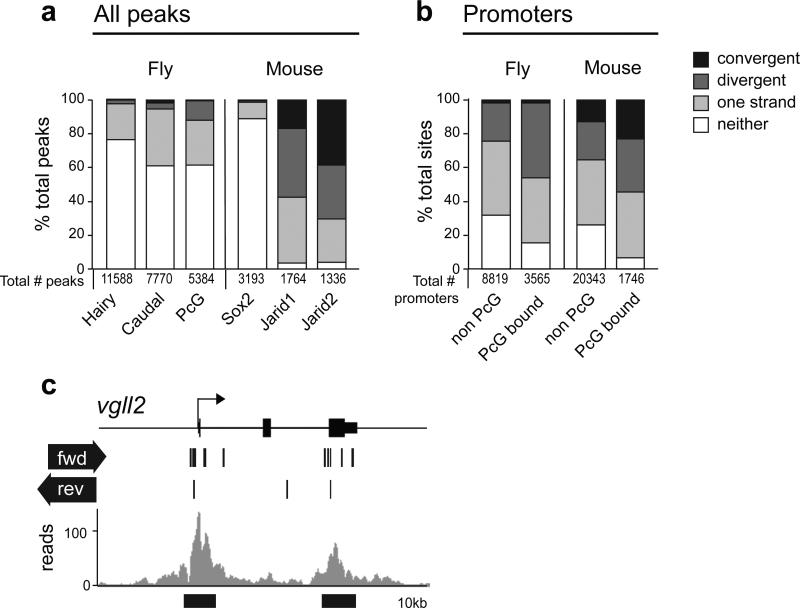
Figure 7. Elements with GEAR box potential are widespread.
(a) Fly: ChIP peaks from21 with CAGE tags from31. Mouse: ChIP peaks from 33,34 compared with CAGE tags from the FANTOM3 CAGE dataset32. Suz12 comparison gave similar results (Supplementary Table 3). % of total peaks overlapping with CAGE tags in the categories shown are indicated. Convergent CAGE tags: tags on opposite strands are transcribed towards each other. Divergent CAGE tags: tags on opposite strands are transcribed away from each other. (b) CAGE tag status of PcG ChIP peaks at promoters was compared to that of non-PcG bound promoters. Fly: data as for (a). Mouse: combined Jarid2 and SUZ12 data from 34 were used for PcG peaks. In most cases, bidirectional transcription occurs significantly more frequently at PcG bound sites than at other sites. The frequency of occurrence of the category of interest (convergent or divergent) compared to all other categories including no transcription, was compared for pairs of data sets (for each comparison, PcG bound was compared to non-PcG bound). p-values (Yates Chi-squared test, one sided). In all cases except caudal, convergent, and Jarid1, divergent, the category of interest was more abundant in PcG than non-PcG bound sites. For fly promoters, convergent, p=0.19; for all other categories, p< 4×106. See also Supplementary Table 2 and Supplementary Table 3, and Methods. (c) Mouse Vgll2 locus: grey, SUZ12 binding in34, black lines, CAGE tag occurrence32. Black bars below plot indicate potential GEAR box elements.