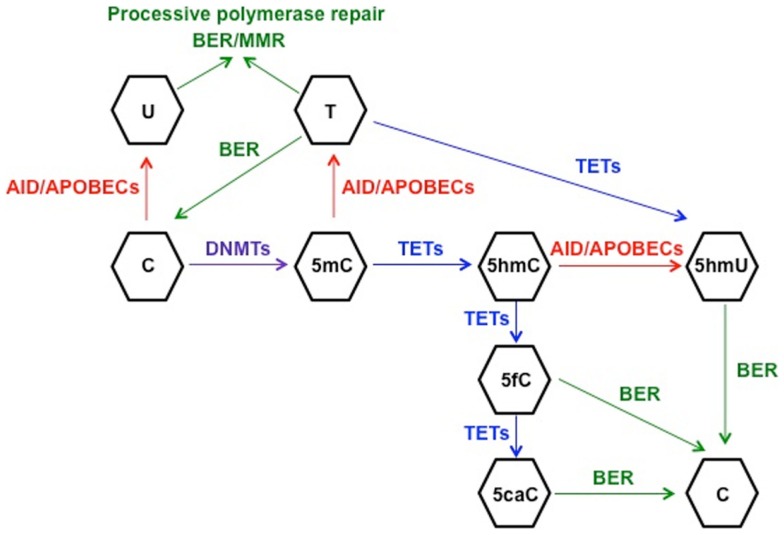
Figure 1.
Schematic representation of the proposed mechanisms for active DNA demethylation. DNMTs catalyze the methylation of cytosine (C) to 5-methyl-C (5mC), which can be deaminated to thymine (T) by AID. The generated mismatch can be repaired by the short-patch BER machinery, restoring C, or by one of the processive repair pathways (long-patch BER or MMR), leading to demethylation of a fragment of DNA. This machinery involving processive DNA polymerases can also repair the uracil:guanine (dU:dG) mismatches generated after C deamination to U. On the other hand, TET-mediated hydroxylation of 5mC generates 5-hydroxymethyl-C (5hmC), which can be deaminated by AID/APOBECs to 5-hydroxymethyl-U (5hmU) and replaced by C through BER. 5hmC can be further oxidized by TETs proteins to 5-formyl-C (5fC) and 5-carboxyl-C (5caC), leading to activation of BER to restore C. Finally, another potential mechanism of active DNA demethylation would involve 5mC deamination to T and TET-mediated oxidation of T to 5hmU, which would be replaced by C through BER, although this model has not been proven as yet.