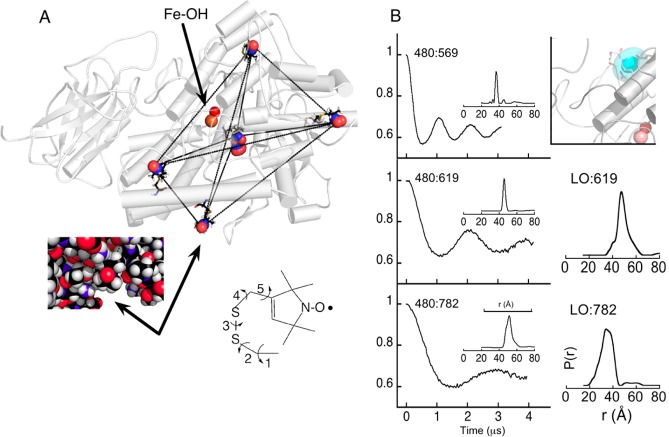
Figure 4.
Lipid polar headgroup located by pulsed dipolar EPR spectroscopy with (A) a grid of five lipoxygenase side chains, mutated in pairs by spin-labeling. The spin-labeled residue numbers are, proceeding clockwise from the uppermost, F270R1, L480R1, A619R1, and F782R1, and the one in the middle is A569R1. The chemical structure shown is the spin-label side chain of R1 that has five rotatable bonds. (B) Calculation of one-dimensional probability distributions, P(r), based on time-domain decays (see Supporting Information in ref (9)). Pairs of residue numbers are indicated, for example, as 480:782 for double-labeled pair L480R1 and F782R1. Structural inset (B, upper right) gives the 1 and 2σ volume distribution solved for the spin of a bound LOPTC (cyan ovals). Pulsed EPR data and analysis were by Peter Borbat (Cornell). Adapted with permission from parts of Figures 2, 3, and 6 of ref (9), 2012, Elsevier.