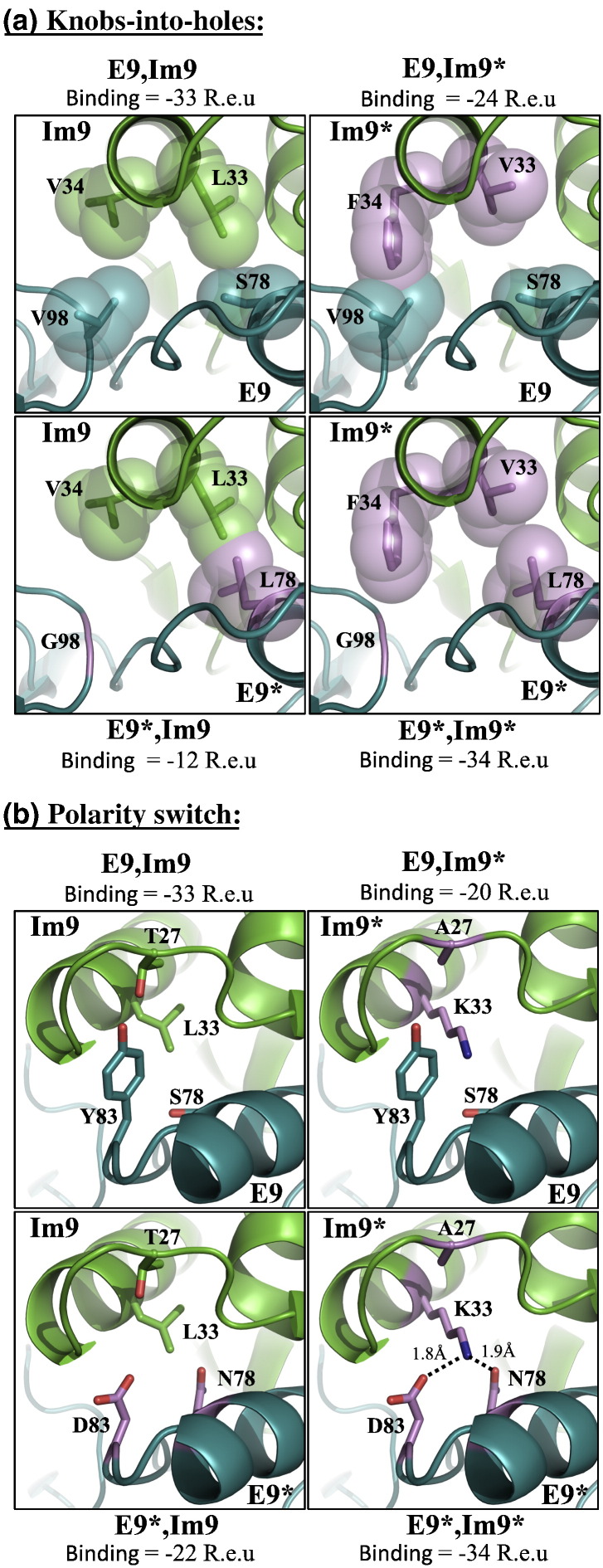
Fig. 5.
Two designed specificity switches in the colicin endonuclease-immunity pair E9-Im9. Interactions with binding energy greater than or equal to − 27 R.e.u. are noncognate (see Results), while interactions with binding energy less than or equal to − 33 are expected to be as favorable as the natural pairs. Im9 and E9 are shown in green and deep-teal cartoons, respectively. The mutated residues in the design pair are shown in violet. (a) A design pair in which specificity is encoded by knobs-into-holes features. In the natural pair (upper left panel), L33 and V34 of Im9 pack against S78 and V98 of E9, respectively. In the design pair (lower right panel), L33V mutation introduces a smaller side chain, and the complementary S78L mutation maintains packing density as in the natural pair. Similarly, V34F introduces a large side-chain in the designed Im9* while V98G removes a side-chain. In the designed noncognate pair E9,Im9*, V34F forms a steric overlap with E9's V98, thus destabilizing the bound state. Similarly, in the designed noncognate pair E9*, Im9 mutation S78L forms a steric overlap with Im9's L33. (b) A design pair, in which specificity is encoded through a switch in polarity features. The natural interaction is mainly based on hydrophobic packing. In the designed pair (bottom right), K33 forms hydrogen bonds with D83 and N78, as well as a salt bridge with D83. In the designed noncognate pairs, the hydrogen bonds are not formed and polar residues pack against hydrophobic ones leading to electrostatic frustration similar to the observation in the natural noncognate pair [74]. Also, there are steric overlaps in the designed noncognate pairs that further destabilize the bound state. In E9*, Im9's (bottom left) D83 and N78 pack against the hydrophobic L33. A steric overlap is formed between L33 and N78. In the design noncognate pair E9,Im9*, K33 packs against S78 and Y83, resulting in steric overlap.