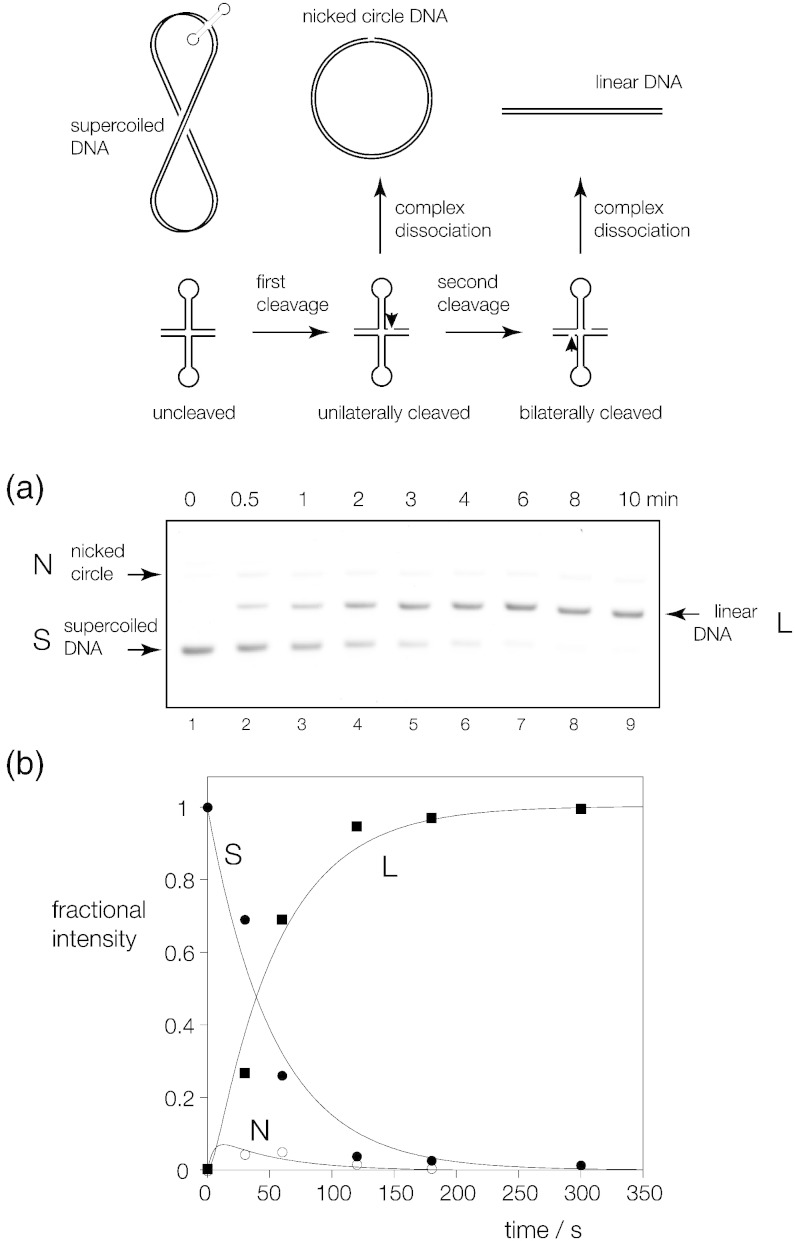
Fig. 7.
Analysis of bilateral cleavage in a DNA junction using a supercoil-stabilized cruciform substrate. The principle of the experiment is shown in the scheme (top). A cruciform structure contains a four-way junction that is a substrate for resolving enzymes. However, the cruciform requires negative supercoiling for stabilization. Unilateral cleavage of the junction followed by dissociation of the protein leads to the formation of a nicked circle in which the cruciform substrate is no longer present. By contrast, subsequent second cleavage within the lifetime of the complex generates a linear product as the cruciform is now bilaterally cleaved.
(a) The supercoiled DNA substrate and the nicked and linear products are readily separated by electrophoresis in 1% agarose. Supercoiled plasmid pBHR3 was incubated with 200 nM CtGEN11–487 in 10 mM Hepes (pH 7.5), 50 mM NaCl, 1 mM DTT and 0.1% BSA at 37 °C. After the cleavage reaction was initiated by addition of MgCl2 to 10 mM, samples were removed at different times, protein was removed by treatment with proteinase K and electrophoresed in a 1% agarose gel and DNA was fluorescently stained. A fluoroimage is shown. With time, the supercoiled DNA is converted to linear product, with a low intensity of nicked circular DNA appearing as a transient intermediate.
(b) The intensities of the three species were quantified and plotted as a function of time (lower). These data are fitted to the integrated rate Eqs. (3), (4), (5) shown in the main text.