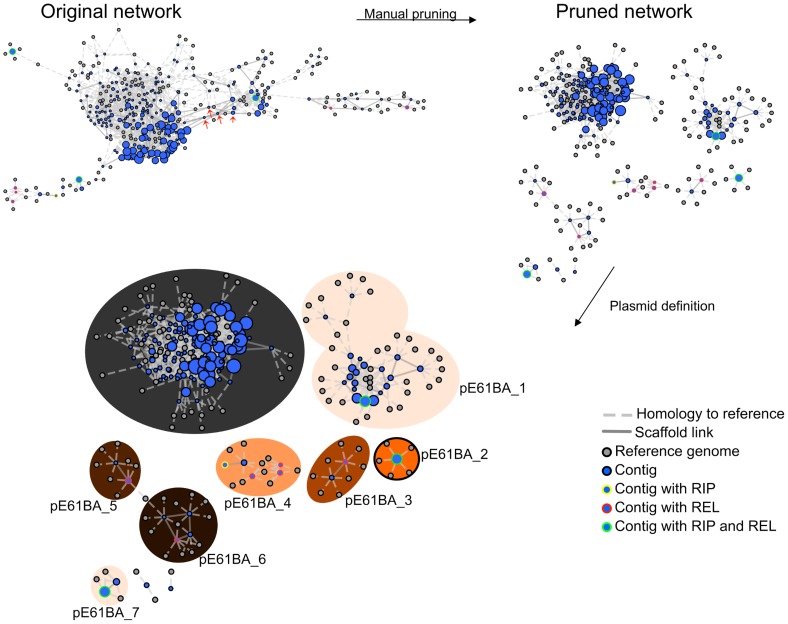
Figure 2. PLACNET plasmid reconstruction of ST131 genome E61BA (ST9/H324/virotype D).
The network contains nodes of two different colors (blue for contigs, grey for reference genomes). The size of reference nodes is always the same. The size of the contig nodes is proportional to the contig length. Besides, outlines are yellow for contigs containing RIP proteins, red for relaxases and green for both proteins. Edges are either solid (scaffold links) of dotted (homologous references). The length of the edges is arbitrarily selected by Cytoscape algorithm. In the upper left, the network output (original network) is shown, which resulted from automatic reference search, scaffold links and protein tagging rules. The original network was converted to a pruned network by eliminating contigs smaller than 200 bp and duplicating specific hubs (red arrows). Two contigs could not be assigned for lack of scaffold links: a 2,953 bp contig (putative DNA primase + lytic transglycosylase) and a 1,301 bp contig (TrbI + TraB-partial). Closed plasmids (e.g., pE61BA_2, size: 24,447 bp) are shown with a black outline in the final PLACNET network.