Abstract
Purpose: This study aimed to evaluate the expression of DNA methyltransferase (DNMT) family proteins in renal cell carcinoma (RCC) and to assess the clinical significance and prognostic value of their expression patterns. Methods: A total of 97 renal cell carcinoma and 52 no-tumor tissues were recruited for immunohistochemical analysis of their expression. Results: DNMT1, DNMT3A and DNMT3B proteins were highly expressed in clear cell RCC, papillary RCC and chromophobe RCC tissues than that of no-tumor tissues (all P < 0.05). DNMT1, DNMT3A and DNMT3B expression was significantly associated with tumor size (P=0.003, 0.001 and 0.003, respectively), tumor pathology stage (P=0.039, 0.034 and 0.037, respectively), histopathological grading (P=0.042, 0.026 and 0.031, respectively), lymph node metastasis (P=0.022, 0.030 and 0.020, respectively) and vascular invasion (P=0.042, 0.031 and 0.044, respectively). The Kaplan-Meier survival analysis demonstrated that expression of DNMTs protein in RCC was significantly associated with shorter over all survival and disease-free survival (all P < 0.05). Furthermore, multivariate analysis showed that the expression of DNMT1 was an independent prognostic factor for overall survival (OS) (P=0.036), and the expression of DNMT3A or DNMT3B was an independent prognostic factor for disease-free survival (DFS) in the patients (P=0.031 and P=0.023, respectively). Conclusions: DNMTs were higher expressed in RCC than no-tumor tissues, and the expression of DNMTs were strongly associated with RCC tumor size, tumor pathology stage, histological grading, lymph node metastasis, vascular invasion, recurrence, and prognosis. DNMTs may thus serve as prognostic markers and novel therapeutic targets for RCC patients.
Keywords: Renal cell carcinoma, DNA methyltransferase, progression, prognosis
Introduction
Renal cell carcinoma (RCC) is a common urologic malignancy and accounts for about 3% of adult malignancies and causes about 90,000 deaths worldwide annually [1]. The rate of RCC has increased by over 2% per year for the past 3 decades [2]. Because RCC has a highly resistant phenotype to conventional therapeutic modalities, including chemotherapy and radiation, surgical resection of localized disease has been regarded as the only curative treatment. However, 20-30% of these patients experience local and/or distant disease recurrence [3]. Moreover, up to 30% of patients have metastases at the time of the initial diagnosis [4]. The prognosis of RCC is very poor, with the highest mortality rate among all the genitourinary tract tumors and a third of patients dying from their disease [5].
Epigenetics refers to stable alterations in gene expression with no underlying modifications in the genetic sequence. Several epigenetic mechanisms regulate gene expression, including DNA methylation, chromatin remodeling, histone variants, and the epigenetic function of non-coding RNA [6]. Among these, DNA methylation is a covalent modification of DNA that plays a significant role in the regulation of gene transcription [7]. Nevertheless, abnormal DNA methylation does also play an important role in human cancer development, and most cancer cells show a global hypomethylation of the genome that induces abnormal expression of genes but a local hypermethylation that silences tumor suppressor genes [6].
Methylation changes to the epigenome are controlled by DNA methyltransferases (DNMTs), which catalyze the transfer of a methyl group from the methyl donor S-adenosyl methionine onto the 5-position on the cytosine ring. Three catalytically active DNMTs have been identified in mammals, DNMT1, DNMT3A, and DNMT3B [8]. The levels of DNMT1, DNMT3A, and DNMT3B mRNA are reportedly elevated in various malignancies, including breast, liver and prostate tumors [9-11].
Similar to other malignancies, aberrant DNA methylation on CpG islands is also an important mechanism for human renal cell carcinoma development. Recently in vitro studies have shown DNMTs inhibitor can induce apoptosis in RCC cells [12]. In addition, another study suggest that DNMTs inhibitor could suppress RCC cell proliferation by inducing G2/M cell cycle arrest and strikingly increase the susceptibility of RCC to paclitaxel [13], leading to using DNMT inhibitors in clinical trials of renal cancers [14-16]. Surprisingly, the specific role of the different DNMT isoforms in carcinogenesis and tumor progression of renal cell carcinoma is not well understood.
In this study, we collected 97 cases of renal cell carcinoma samples and 52 cases of adjacent non-tumor tissues for detection of DNMT family protein expressions in order to determine the role of DNMT proteins in renal cancer and clinical significance.
Patients and tissue specimens
Renal cell carcinoma tissue was collected from radical nephretomy specimens performed between January 2004 and January 2012 at Department of Urology, Shengjing Hospital of China Medical University. The tumor cases included 97 cases with histologically confirmed malignant carcinoma and 52 cases of adjacent non-tumor tissues. The criteria for study enrollment were as follows: patients with histopathologically diagnosed RCC who were newly diagnosed, untreated without a history of other tumors, and subsequently underwent radical nephrectomy. Histological diagnosis was established according to the guidelines of the World Health Organization [17]. Cases were selected according to tissue availability and were not stratified for any known preoperative or prognostic factor. None of the patients underwent chemotherapy, radiotherapy, or adjuvant treatment before surgery. We obtained the written informed consent from all the patients. The Institutional Review Board of China Medical University approved the research protocol. The patients were carefully followed up by consulting their case documents and through telephone monitoring.
Immunohistochemistry
Formalin-fixed and paraffin-embedded tissue samples were cut into 4-mm thick sections and mounted onto poly-L-lysine-coated glass slides. For immunohistochemical staining, the sections were deparaffinized in xylene, rehydrated in a series of alcohol, and washed in the tap water. The sections were then cooked in 10 mM sodium citrate buffer, pH 6.0, for 10 min in an autoclave for antigen retrieval. Endogenous peroxidase activity was blocked by incubating the sections in 3% H2O2 at 37°C for 20 min. After that, the sections were blocked to avoid nonspecific binding by addition of a 10% normal goat serum at 37°C for 30 min and then incubated for 4°C overnight with the polyclonal antibody against DNMTs (DNMT1, sc-20701, 1:250 dilution; DNMT3a, sc-20703, 1:250 dilution; DNMT3b sc-130740, 1:250 dilution; Santa Cruz Biotechnology, USA). The specificity of antibodies had been confirmed by using Western blot analysis (data not shown). In the next day, the sections were washed five times with 0.01 mol/L phosphate-buffered saline (PBS; pH 7.4) for 15 min and then incubated with a biotinylated secondary antibody for 30 min at 37°C in the dark. After that, the sections were incubated with a streptavidin horseradish peroxidase solution for another 30 min (LSAB kit; Dako, Glostrup, Denmark), washed in PBS, and stained with DAB (3, 3-diaminobenzidine). Finally, the sections were counterstained with Mayer’s hematoxylin, dehydrated, and mounted. Negative controls were run in parallel, and were generated by PBS replacing the anti-DNMTs antibody.
Evaluation of immunohistochemistry
The immunostained sections were evaluated by two investigators who were blinded to the patients’ clinicopathological characteristics. For each slide, the number of DNMTs positive cells was counted in 10 fields at ×200 magnification, and the percentage of positively stained cells was determined. The percentage of positively stained cells was graded semi-quantitatively according to a four-point scoring system as follows: negative (-), 0; weakly positive (+), < 25%; moderately positive (++), 26-50%; and strongly positive (+++), > 50%.
Statistical analysis
Statistical analyses were performed with SPSS 19.0 (SPSS Inc., Chicago, USA). Comparison of DNMTs expression between samples was analyzed by using the Mann-Whitney U-test. Chi-square tests were applied to assess associations between expression of DNMTs and clinicopathological parameters. Univariate survival analysis was carried out according to Kaplan-Meier, differences in survival curves were assessed with the log rank test. Cox regression analysis was used for the multivariate analysis. P-values < 0.05 were considered significant.
Results
Patient characteristics
The clinicopathological data from the patients are shown in Table 1. The mean age of the patients at surgery was 53 years (rang 15-84), and 59 (60.8%) of the patients were diagnosed before 55 years old. 60 (61.9%) were male. 60 (61.9%) patients had clear cell RCC (ccRCC), 23 (23.7%) papillary RCC (pRCC) and 14 (14.4%) chromophobe RCC (chRCC). Clinical follow-up data, as annually assessed survival time was available for all patients. The median follow-up time of all cases was 50 months, ranging from 12 to 118 months. 38 (39.2%) patients exhibited recurrence and 29 (29.9%) patients died from renal cancer during follow up. The pT status was as follows: pT1a-20 (20.6%), pT1b-25 (25.8%), pT2a-17 (17.5%), pT2b-11 (11.3%), pT3a-16 (16.5%), pT3b-2 (2.1%), and pT4-6 (6.2%). 14 patients (14.4%) had pathologically confirmed nodal metastases. 62 (63.9%) patients had no nodal metastases (pN0). In 21 (21.6%) patients lymph nodes were not examined (pNx). Tumor grades, according to Fuhrman, were G1-25 (25.8%), G2-33 (34%), G3-27 (27.8%) and G4-12 (12.4%), respectively.
Table 1.
Patient characteristics
| Feature | Categories | Number | % |
|---|---|---|---|
| Gender | |||
| Female | 37 | 38.1 | |
| Male | 60 | 61.9 | |
| Age, years | |||
| ≤ 55 | 59 | 60.8 | |
| > 55 | 38 | 39.2 | |
| Histological type | |||
| Clear cell RCC | 60 | 61.9 | |
| Papillary RCC | 23 | 23.7 | |
| Type1 | 10 | 10.3 | |
| Type2 | 13 | 13.4 | |
| Chomophobe RCC | 14 | 14.4 | |
| Tumor size (cm) | |||
| ≤ 4 | 20 | 20.6 | |
| > 4 | 77 | 79.4 | |
| Tumor pathology stage | |||
| pT1a | 20 | 20.6 | |
| pT1b | 25 | 25.8 | |
| pT2a | 17 | 17.5 | |
| pT2b | 11 | 11.3 | |
| pT3a | 16 | 16.5 | |
| pT3b | 2 | 2.1 | |
| pT4 | 6 | 6.2 | |
| Grading | |||
| G1 | 25 | 25.8 | |
| G2 | 33 | 34.0 | |
| G3 | 27 | 27.8 | |
| G4 | 12 | 12.4 | |
| Lymph node metastasis | |||
| pNx | 21 | 21.6 | |
| pN0 | 62 | 63.9 | |
| pN1/2 | 14 | 14.4 | |
| Vascular invasion | |||
| No | 91 | 93.8 | |
| Yes | 6 | 6.2 | |
| Tobacco smoking | |||
| No | 41 | 42.3 | |
| yes | 56 | 57.7 |
Expression of DNMTs in renal cell cancer and no-tumor tissues
Differential expression of DNMT proteins in renal cell carcinoma and no-tumor tissues according to histological type is summarized in Table 2. In our study, DNMT proteins were significantly highly expressed in different subtypes of renal cell cancer tissues than that of no-tumor tissues (Mann-Whitney U-test, all P < 0.05). Briefly, The positive rates for DNMT1, DNMT3A, and DNMT3B expression in the ccRCC tissues were 56.7%, 63.3%, and 65.0%, respectively, which were significantly higher than those of no-tumor tissues (27.3, 31.8%, and 36.4%, respectively); the positive rates for DNMT1, DNMT3A, and DNMT3B expression in the pRCC tissues were 60.9%, 65.2%, and 69.6%, respectively, which were significantly higher than those of no-tumor tissues (27.8%, 27.8%, and 22.2%, respectively); and the positive rates for DNMT1, DNMT3A, and DNMT3B expression in the chRCC tissues were 64.3%, 78.6%, and 78.6%, respectively, which were significantly higher than those of no-tumor tissues (16.7%, 33.3%, and 25.0%, respectively). Representative expression patterns of immunohistochemical staining of DNMTs in RCC and non-tumor tissues were shown in Figures 1, 2 and 3, respectively.
Table 2.
DNMTs expression in RCC and non-tumor tissues
| - | + | ++ | +++ | |||||
|---|---|---|---|---|---|---|---|---|
|
|
||||||||
| n | N (%) | N (%) | N (%) | N (%) | PR, % | P-value | ||
| Clear cell RCC | DNMT1 | |||||||
| Tumor | 60 | 26 (43.3) | 22 (36.7) | 9 (15.0) | 3 (5.0) | 56.7 | 0.019 | |
| non-Tumor | 22 | 16 (72.7) | 3 (13.6) | 2 (9.1) | 1 (4.5) | 27.3 | ||
| DNMT3A | ||||||||
| Tumor | 60 | 22 (36.7) | 20 (33.3) | 13 (21.7) | 5 (8.3) | 63.3 | 0.012 | |
| non-Tumor | 22 | 15 (68.2) | 4 (18.2) | 2 (9.1) | 1 (4.5) | 31.8 | ||
| DNMT3B | ||||||||
| Tumor | 60 | 21 (35.0) | 24 (40.0) | 11 (18.3) | 4 (6.7) | 65.0 | 0.021 | |
| non-Tumor | 22 | 14 (63.6) | 4 (18.2) | 3 (13.6) | 1 (4.5) | 36.4 | ||
| Papillary RCC | DNMT1 | |||||||
| Tumor | 23 | 9 (39.1) | 7 (30.4) | 4 (17.4) | 3 (13.0) | 60.9 | 0.037 | |
| non-Tumor | 18 | 13 (72.2) | 2 (11.1) | 2 (11.1) | 1 (5.6) | 27.8 | ||
| DNMT3A | ||||||||
| Tumor | 23 | 8 (34.8) | 7 (30.4) | 5 (21.7) | 3 (13.0) | 65.2 | 0.019 | |
| non-Tumor | 18 | 13 (72.2) | 3 (16.7) | 2 (11.1) | 0 (0) | 27.8 | ||
| DNMT3B | ||||||||
| Tumor | 23 | 7 (30.4) | 6 (26.1) | 7 (30.4) | 3 (13.0) | 69.6 | 0.003 | |
| non-Tumor | 18 | 14 (77.8) | 3 (16.7) | 1 (5.6) | 0 (0) | 22.2 | ||
| Chomophobe RCC | DNMT1 | |||||||
| Tumor | 14 | 5 (35.7) | 4 (28.6) | 3 (21.4) | 2 (14.3) | 64.3 | 0.041 | |
| non-Tumor | 12 | 10 (83.3) | 1 (8.3) | 1 (8.3) | 0 (0) | 16.7 | ||
| DNMT3A | ||||||||
| Tumor | 14 | 3 (21.4) | 5 (35.7) | 5 (35.7) | 1 (7.1) | 78.6 | 0.022 | |
| non-Tumor | 12 | 8 (66.7) | 2 (16.7) | 1 (8.3) | 1 (8.3) | 33.3 | ||
| DNMT3B | ||||||||
| Tumor | 14 | 3 (21.4) | 6 (42.9) | 3 (21.4) | 2 (14.3) | 78.6 | 0.007 | |
| non-Tumor | 12 | 9 (75.0) | 2 (16.7) | 1 (8.3) | 0 (0) | 25.0 | ||
-, negative; +, weak; ++, moderate; +++, strong staining; PR, positive rate. P-value obtained from Mann-Whitney U test.
Figure 1.
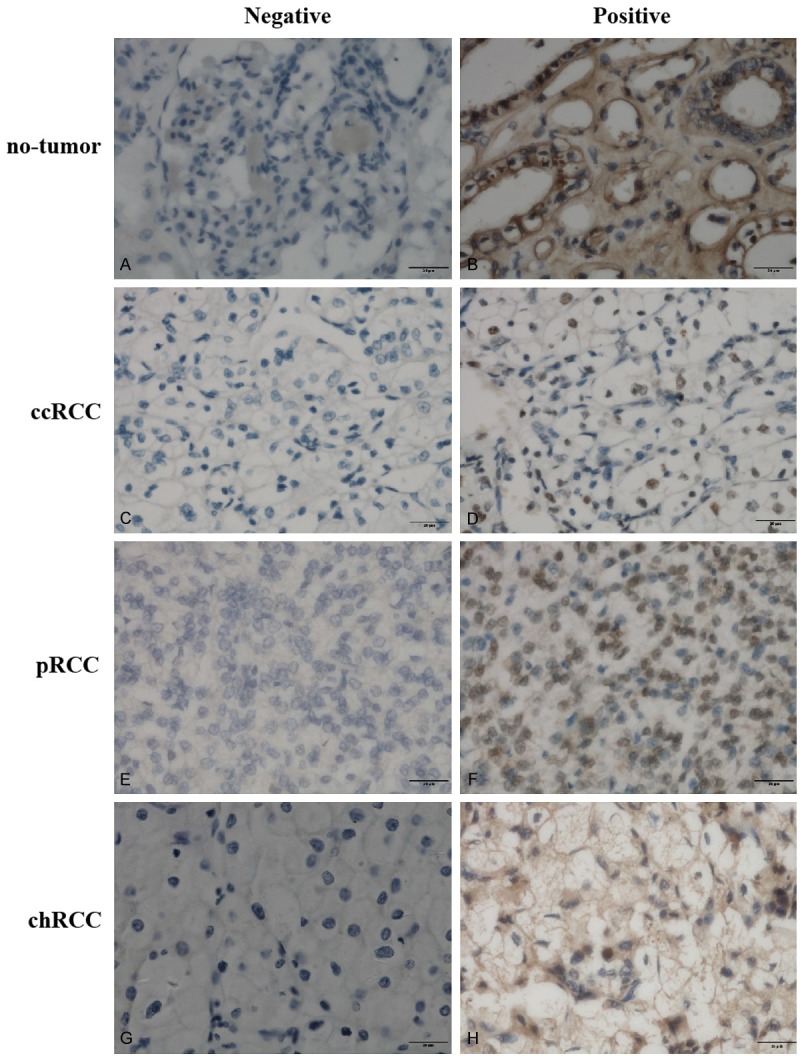
Representative immunohistochemical staining of DNMT1 protein in tissue samples (original magnification, ×400 in A-H).
Figure 2.
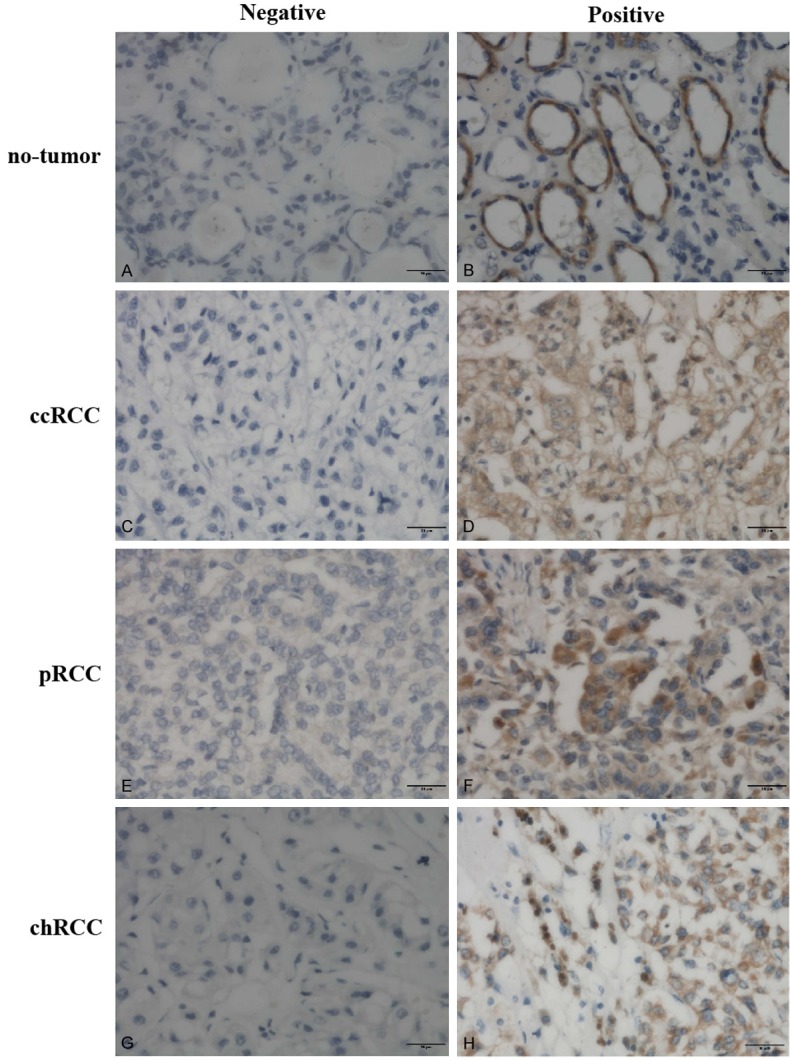
Representative immunohistochemical staining of DNMT3A protein in tissue samples (original magnification, ×400 in A-H).
Figure 3.
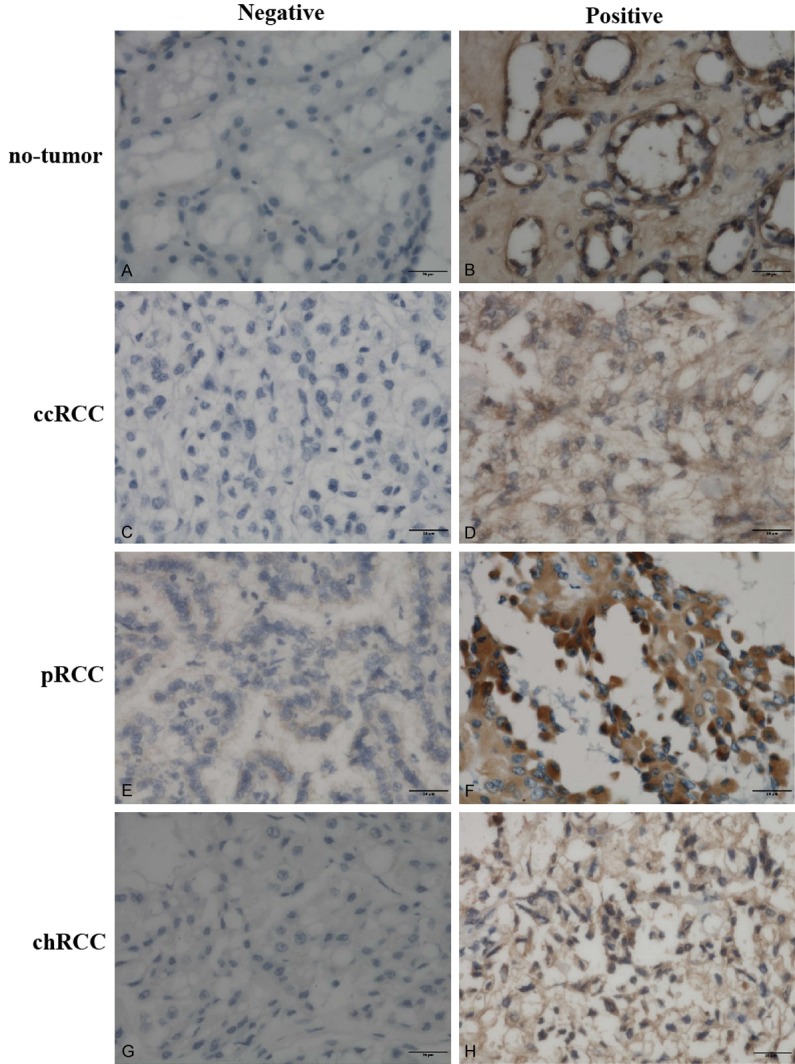
Representative immunohistochemical staining of DNMT3B protein in tissue samples (original magnification, ×400 in A-H).
Association between expression of DNMT proteins and clinicopathological parameters
The correlation analysis of DNMTs protein with clinicopathological factors in RCC was shown in Table 3. Our data showed that DNMT1, DNMT3A and DNMT3B expression was significantly associated with tumor size (P=0.003, 0.001 and 0.003, respectively), tumor pathology stage (P=0.039, 0.034 and 0.037, respectively), histopathological grading (P=0.042, 0.026 and 0.031, respectively), lymph node metastasis (P=0.022, 0.030 and 0.020, respectively), and vascular invasion (P=0.042, 0.031 and 0.044, respectively). However, there were no association between DNMT1, DNMT 3A and DNMT3B expression with gender (P=0.244, 0.533 and 0.155, respectively), age (P=0.889, 0.975 and 0.949, respectively), or tobacco smoking (P=0.481, 0.095 and 0.980, respectively).
Table 3.
Correlation between DNMTs expression and clinicopathological factors of RCC
| Parameter | n | DNMT1 n (%)a | DNMT3a n (%) | DNMT3b n (%) | |
|---|---|---|---|---|---|
| Gender | |||||
| Female | 37 | 19 (51.4) | 23 (62.2) | 22 (59.5) | |
| Male | 60 | 38 (63.3) | 41 (68.3) | 44 (73.3) | |
| P b | 0.244 | 0.533 | 0.155 | ||
| Age at diagnosis | |||||
| ≤ 55 | 59 | 35 (59.3) | 39 (66.1) | 40 (67.8) | |
| > 55 | 38 | 22 (57.9) | 25 (65.8) | 26 (68.4) | |
| P | 0.889 | 0.975 | 0.949 | ||
| Tumor size | |||||
| ≤ 4 | 20 | 6 (30.0) | 7 (35.0) | 8 (40.0) | |
| > 4 | 77 | 51 (66.2) | 57 (74.0) | 58 (75.3) | |
| P | 0.003 | 0.001 | 0.003 | ||
| Tumor pathology stage | |||||
| pT1 | 45 | 20 (44.4) | 23 (51.1) | 24 (53.3) | |
| pT2 | 28 | 18 (64.3) | 21 (75.0) | 22 (78.6) | |
| pT3 | 18 | 14 (77.8) | 15 (83.3) | 15 (83.3) | |
| pT4 | 6 | 5 (83.3) | 5 (83.3) | 5 (83.3) | |
| P | 0.039 | 0.034 | 0.037 | ||
| Grading | |||||
| G1 | 25 | 9 (36.0) | 12 (48.0) | 13 (52.0) | |
| G2 | 33 | 20 (60.6) | 20 (60.6) | 21 (63.6) | |
| G3 | 27 | 19 (70.4) | 21 (77.8) | 21 (77.8) | |
| G4 | 12 | 9 (75.0) | 11 (91.7) | 11 (91.7) | |
| P | 0.042 | 0.026 | 0.031 | ||
| Lymph node metastasis | |||||
| pNx | 21 | 16 (76.2) | 17 (81.0) | 18 (85.7) | |
| pN0 | 62 | 30 (48.4) | 35 (56.5) | 36 (58.1) | |
| pN1/2 | 14 | 11 (78.6) | 12 (85.7) | 12 (85.7) | |
| P | 0.022 | 0.030 | 0.020 | ||
| Vascular invasion | |||||
| No | 84 | 46 (54.8) | 52 (61.9) | 54 (64.3) | |
| Yes | 13 | 11 (84.6) | 12 (92.3) | 12 (92.3) | |
| 0.042 | 0.031 | 0.044 | |||
| Tobacco smoking | |||||
| No | 69 | 39 (56.5) | 42 (60.9) | 47 (68.1) | |
| yes | 28 | 18 (64.3) | 22 (78.6) | 19 (67.9) | |
| P | 0.481 | 0.095 | 0.980 |
Numbers in parentheses are percentage.
P-value obtained from Chi-Square test.
Association of DNMT protein expressions with survival of the patients
The correlation between DNMTs protein expression and prognosis in RCC patients was analyzed with the Kaplan-Meier method. We observed that the expression of DNMT1, DNMT3A and DNMT3B proteins in RCC was significantly correlated with DFS and OS (all P < 0.05; Table 4). The log-rank test further demonstrated that the OS and DFS time were both significantly different between groups with and without expression of DNMTs protein, which indicated expression of DNMTs protein was correlated with a shorter survival time (Figure 4). Other clinicopathologic parameters, including histological type (P=0.003 and 0.026, respectively), tumor size (P=0.006 and 0.021, respectively), tumor pathology stage (P=0.000 and 0.000, respectively), histological grade (P=0.024 and 0.004, respectively), lymph node metastasis (P=0.000 and 0.001, respectively), and vascular invasion (P=0.019 and 0.006, respectively) were also significantly correlated with overall survival and disease-free survivals in univariate analysis (Table 4). In addition, multivariate analysis using the Cox proportional hazards model showed that the expression of DNMT1 was an independent prognostic factor for over all survival (P=0.036), and the DNMT3A or DNMT3B expression were independent predictors of disease-free survival in patients with RCC (P=0.031 and 0.023, respectively), the traditional tumor size, tumor pathology stage, lymph node metastasis, and vascular invasion were independent predictors of both over all survival and disease-free survival (all P < 0.05) (Table 5).
Table 4.
Univariate analysis of OS and DFS in patients with RCC (months, mean ± SE)
| Variable | n | Overall survival time | P | Disease-free survival time | P |
|---|---|---|---|---|---|
| Gender | |||||
| Female | 37 | 70.527±5.960 | 0.461 | 67.001±6.542 | 0.811 |
| Male | 60 | 84.772±6.662 | 73.614±6.481 | ||
| Age, years | |||||
| ≤ 55 | 59 | 83.286±6.505 | 0.810 | 75.783±6.511 | 0.709 |
| > 55 | 38 | 72.984±5.755 | 63.374±6.242 | ||
| Histological type | |||||
| Clear cell RCC | 60 | 82.807±6.102 | 0.003 | 74.754±6.219 | 0.026 |
| Papillary RCC | 23 | 58.625±8.444 | 53.610±8.642 | ||
| Chomophobe RCC | 14 | 92.333±7.893 | 82.833±8.982 | ||
| Tumor size (cm) | |||||
| ≤ 4 | 20 | 104.778±7.894 | 0.006 | 91.703±8.859 | 0.021 |
| > 4 | 77 | 67.498±4.322 | 61.987±4.609 | ||
| Tumor pathology stage | |||||
| pT1a | 20 | 96.489±8.586 | 0.000 | 83.074±9.445 | 0.000 |
| pT1b | 25 | 81.086±6.106 | 73.404±6.367 | ||
| pT2a | 17 | 70.261±7.119 | 55.833±6.716 | ||
| pT2b | 11 | 57.659±5.723 | 52.773±5.175 | ||
| pT3a | 16 | 50.500±4.450 | 45.269±4.621 | ||
| pT3b | 2 | 48.000±8.000 | 29.500±0.500 | ||
| pT4 | 6 | 28.500±5.374 | 24.167±7.295 | ||
| Grading | |||||
| G1 | 25 | 83.484±9.098 | 0.024 | 77.446±9.586 | 0.004 |
| G2 | 33 | 80.976±5.847 | 75.247±6.307 | ||
| G3 | 27 | 70.065±7.626 | 61.312±7.579 | ||
| G4 | 12 | 37.661±4.288 | 34.333±4.667 | ||
| Lymph node metastasis | |||||
| pNx | 21 | 72.742±7.890 | 0.000 | 68.129±8.470 | 0.001 |
| pN0 | 62 | 89.848±6.287 | 78.247±6.116 | ||
| pN1/2 | 14 | 47.449±6.461 | 32.643±4.840 | ||
| Vascular invasion | |||||
| No | 91 | 84.366±5.300 | 0.019 | 75.496±5.276 | 0.006 |
| Yes | 6 | 48.625±10.085 | 31.333±4.148 | ||
| Tobacco smoking | |||||
| No | 41 | 91.782±6.990 | 0.082 | 76.597±7.049 | 0.440 |
| yes | 56 | 66.319±4.705 | 63.857±5.480 | ||
| DNMT1 expression | |||||
| Positive | 57 | 61.117±4.768 | 0.004 | 82.823±6.375 | 0.004 |
| Negative | 40 | 92.260±6.174 | 57.251±5.098 | ||
| DNMT3A expression | |||||
| Positive | 64 | 73.291±8.061 | 0.016 | 68.915±8.213 | 0.034 |
| Negative | 33 | 85.291±4.383 | 76.700±5.095 | ||
| DNMT3B expression | |||||
| Positive | 66 | 71.837±7.326 | 0.011 | 63.623±7.690 | 0.003 |
| Negative | 31 | 87.223±4.534 | 80.382±5.157 |
Figure 4.
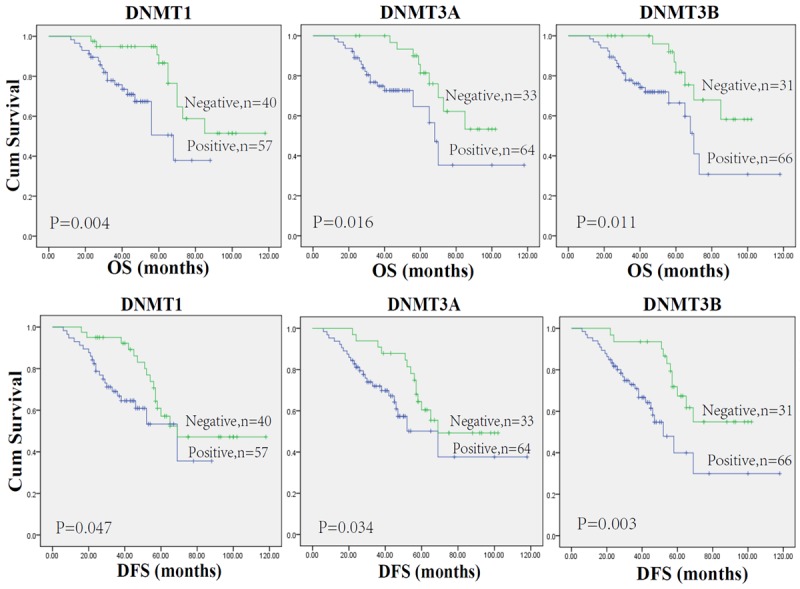
Kaplan-Meier estimates of patients with renal cell carcinoma stratified by DNMTs protein expression.
Table 5.
Multivariate analysis of prognostic factors in patients with RCC
| Factor | Overall survival | Disease-free survival | ||||
|---|---|---|---|---|---|---|
|
|
|
|||||
| RRa | 95% CIb | P | RRa | 95% CIb | P | |
| Gender | ||||||
| Male/Female | 1.083 | 0.672-2.817 | 0.853 | 1.214 | 0.713-2.901 | 0.879 |
| Age, years | ||||||
| > 55/≤ 55 | 1.348 | 0.621-2.674 | 0.506 | 1.268 | 0.603-2.593 | 0.623 |
| Tumor size, cm | ||||||
| ≤ 4/> 4 | 7.325 | 4.267-12.383 | 0.031 | 7.865 | 4.612-13.013 | 0.019 |
| Tumor pathology stage | ||||||
| T3-T4/T1-2 | 8.455 | 5.216-13.478 | 0.017 | 8.633 | 5.341-13.652 | 0.014 |
| Grading | ||||||
| 3, 4/1, 2 | 3.231 | 2.216-6.485 | 0.022 | 2.975 | 2.014-3.880 | 0.015 |
| Lymph node metastasis | ||||||
| yes/no | 3.452 | 2.421-6.533 | 0.012 | 3.623 | 2.583-6.632 | 0.008 |
| Vascular invasion | ||||||
| yes/no | 2.286 | 1.783-5.043 | 0.017 | 2.632 | 1.937-5.232 | 0.011 |
| Tobacco smoking | ||||||
| yes/no | 1.476 | 0.843-3.246 | 0.683 | 1.547 | 0.891-3.359 | 0.579 |
| DNMT1 status | ||||||
| positive/negative | 1.598 | 0.926-2.021 | 0.036 | 1.683 | 1.053-2.416 | 0.121 |
| DNMT3A status | ||||||
| positive/negative | 1.662 | 0.985-2.376 | 0.208 | 1.717 | 1.097-2.502 | 0.031 |
| DNMT3B status | ||||||
| positive/negative | 1.713 | 1.035-2.681 | 0.192 | 1.845 | 1.106-2.583 | 0.023 |
RR, relative risk;
95% CI, 95% confidence interval.
Discussion
In the present study, we immunohistochemically determined the expression of DNMT family proteins in RCC and adjacent non-tumor tissues, additionally, we analyzed the association with expression of DNMTs protein to the survival. The data showed that DNMT1 family protein expression was higher in the subtype of ccRCC, pRCC and chRCC than that in adjacent non-tumor tissues, which was consistent with the previous studies in other types of cancer [10,18-20]. Until now, there is minimal amount of report on DNMT protein in RCC. A report in 2006 [21] found that the incidence of nuclear immunoreactivity for DNMT1 tended to be higher in proximal tubules from nontumorous renal tissues than in those from normal renal tissues, and was significantly higher in ccRCCs. However, there were no DNMT3A and DNMT3B. In our study, we revealed for the first time that the expression of DNMT1, DNMT3A and DNMT3B are highly expressed in ccRCC, pRCC and chRCC, and demonstrated that the expression of DNMT proteins is associated with a more aggressive behavior of RCC. In addition, we validated the expression of DNMTs was the independent risk factor of prognosis for RCC. Collecting duct RCC was a significant and important group of epithelial tumor of the RCC. We have also collected collecting duct RCC but the number of collecting duct RCC available in our study were only 2, so we do not analyzed the collecting duct RCC in this study, and we will continue to collect more collecting duct RCC for future study.
Our current study further associated the relevance of DNMT family protein expressions with clinicopathological features from RCC patients. We found that the DNMTs protein expression was positively linked to tumor size, pathological stage, histological grade, lymph node metastasis, and vascular invasion in RCC. Moreover, in our univariate analysis showed that the expression of DNMT1, DNMT3A or DNMT3B protein, tumor size, histological type, tumor pathology stage, lymph node metastasis, and vascular invasion were associated with prognosis in RCC patients. The Overall survival (OS) and disease-free survival (DFS) of RCC patients were worse with expression of DNMT1, DNMT3A or DNMT3B compared with the patients with out expression of DNMT proteins. In addition, our multivariate analysis showed that the expression of DNMT1 was an independent prognostic factor of OS, and the expression of DNMT3A or DNMT3B was independent risk factor of DSF for RCC patients. In a variety of cancers, DNMT1, DNMT3A, and DNMT3B were reported to be highly expressed and associated with poor prognosis, including bladder, prostate, breast, lung, liver, cervical cancers [9,10,18,22-24]. The prognostic association between DNMT family and RCC patients may be because of the function role of “maintenance methylation” of DNMT1 and the “de nove methylation” of DNMT3A and DNMT3B.
Hypermethylation of some tumor suppress gene (TSG) promoters that affect the prognosis can also explain the poor prognosis associated with patients with DNMT overexpression [25-27]. In RCC, a series of investigations about the identification of methylated tumor suppressor gene candidates have been carried out. The tumor suppressor genes silenced by methylation in RCC include RASSF1A [28], VHL [29]. In addition, a recent report demonstrated that BNC1, PDLIM4, RPRM, CST6, SFRP1, GREM1, COL14A1 and COL15A1 showed frequent tumor-specific promoter region methylation and hypermethylation was associated with transcriptional silencing of TSG [30]. DNA methylation and histone modifications are intricately connected with each other. Histone deacetylase may play an important role in cooperation with DNA methyltransferases, in maintaining tumor suppressor gene silencing [31]. Thus, the use of a combination of inhibitors of DNMTs and histone deacetylase is an attractive therapeutic strategy in cancer [32]. In addition, microRNAs are also involved in regulation of DNMT expression [33,34]. Recent evidence showed that microRNA (miRs), which are noncoding RNAs, can be involved in the promoter methylation of CpG islands by targeting DNMTs 3’UTR [33-35]. In any events, due to the complex mechanisms responsible for regulation of DNMT expressions and functions of DNMTs in carcinogenesis, the altered expression and effects of DNMTs should be further investigated in RCC.
The development of molecular biomarkers is clinically important in that they can help identifying tumors and predicting the recurrence and progression of tumors, which could potentially be benefit for effective treatments. Our data suggested that the expression of DNMT proteins is correlated with more aggressive tumors. Moreover, DNMTs could potentially be added to the current prognostic methods to improve the accuracy of clinical outcome predictions and the choice of appropriate therapy for RCC patients who might have disease recurrence and progression. The findings are potentially important in practice, because the expression of DNMTs could be a potential indicator for the use of DNMT inhibtor in the treatment of metastatic RCC.
In summary, we demonstrated that DNMT family proteins were higher expressed in RCC than no-tumor tissues, and the expression of DNMTs were strongly associated with RCC tumor size, tumor stage, histological grading, lymph node metastasis, vascular invasion, recurrence, and prognosis. DNMTs may thus serve as prognostic markers and novel therapeutic targets for RCC patients. However, this was a retrospective cohort research, so further multicenter studies are needed to confirm these findings and provide more evidence about the role and significance of DNMTs in renal cell carcinoma.
Acknowledgements
This work was supported by grants from National Natural Science Foundation of China (No. 30873097). We thank Shanye Yin for his excellent language editing.
Disclosure of conflict of interest
None.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008. GLOBOCAN 2008. Int J Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 3.Rini BI, Rathmell WK, Godley P. Renal cell carcinoma. Curr Opin Oncol. 2008;20:300–6. doi: 10.1097/CCO.0b013e3282f9782b. [DOI] [PubMed] [Google Scholar]
- 4.Janzen NK, Kim HL, Figlin RA, Belldegrun AS. Surveillance after radical or partial nephrectomy for localized renal cell carcinoma and management of recurrent disease. Urol Clin North Am. 2003;30:843–52. doi: 10.1016/s0094-0143(03)00056-9. [DOI] [PubMed] [Google Scholar]
- 5.O’Rourke CJ, Knabben V, Bolton E, Moran D, Lynch T, Hollywood D, Perry AS. Manipulating the epigenome for the treatment of urological malignancies. Pharmacol Ther. 2013;138:185–196. doi: 10.1016/j.pharmthera.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Matouk CC, Marsden PA. Epigenetic regulation of vascular endothelial gene expression. Circ Res. 2008;102:873–887. doi: 10.1161/CIRCRESAHA.107.171025. [DOI] [PubMed] [Google Scholar]
- 8.Jeltsch A. Beyond Watson and Crick: DNA methylation and molecular enzymology of DNA methyltransferases. Chembiochem. 2002;3:274–293. doi: 10.1002/1439-7633(20020402)3:4<274::AID-CBIC274>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 9.Girault I, Tozlu S, Lidereau R, Bieche I. Expression analysis of DNA methyltransferases 1, 3A, and 3B in sporadic breast carcinomas. Clin Cancer Res. 2003;9:4415–4422. [PubMed] [Google Scholar]
- 10.Saito Y, Kanai Y, Nakagawa T, Sakamoto M, Saito H, Ishii H, Hirohashi S. Increased protein expression of DNA methyltransferase (DNMT) 1 is significantly correlated with the malignant potential and poor prognosis of human hepatocellular carcinomas. Int J Cancer. 2003;105:527–532. doi: 10.1002/ijc.11127. [DOI] [PubMed] [Google Scholar]
- 11.Patra SK, Patra A, Zhao H, Dahiya R. DNA methyltransferase and demethylase in human prostate cancer. Mol Carcinog. 2002;33:163–171. doi: 10.1002/mc.10033. [DOI] [PubMed] [Google Scholar]
- 12.Konac E, Varol N, Yilmaz A, Menevse S, Sozen S. DNA methyltransferase inhibitor mediated apoptosis in the Wnt β catenin signal pathway in a renal cell carcinoma cell line. Exp Biol Med. 2013;238:1009–1016. doi: 10.1177/1535370213498984. [DOI] [PubMed] [Google Scholar]
- 13.Shang D, Ito N, Kamoto T, Ogawa O. Demethylating agent 5-aza-2’-deoxycytidine enhances susceptibility of renal cell carcinoma to paclitaxel. Urology. 2007;69:1007–1012. doi: 10.1016/j.urology.2007.02.026. [DOI] [PubMed] [Google Scholar]
- 14.Stewart DJ, Donehower RC, Eisenhauer EA, Wainman N, Shah AK, Bonfils C, MacLeod AR, Besterman JM, Reid GK. A phase I pharmacokinetic and pharmacodynamic study of the DNA methyltransferase 1 inhibitor MG98 administered twice weekly. Ann Oncol. 2003;14:766–774. doi: 10.1093/annonc/mdg216. [DOI] [PubMed] [Google Scholar]
- 15.Winquist E, Knox J, Ayoub JP, Wood L, Wainman N, Reid GK, Pearce L, Shah A, Eisenhauer E. Phase II trial of DNA methyltransferase 1 inhibition with the antisense oligonucleotide MG98 in patients with metastatic renal carcinoma, a National Cancer Institute of Canada Clinical Trials Group investigational new drug study. Invest New Drugs. 2006;24:159–167. doi: 10.1007/s10637-006-5938-1. [DOI] [PubMed] [Google Scholar]
- 16.Amato RJ, Stephenson J, Hotte S, Nemunaitis J, Bélanger K, Reid G, Martell RE. MG98, a second-generation DNMT1 inhibitor, in the treatment of advanced renal cell carcinoma. Cancer Invest. 2012;30:415–421. doi: 10.3109/07357907.2012.675381. [DOI] [PubMed] [Google Scholar]
- 17.Eble JN, Sauter G, Epstein JI, Sesterhenn IA. Pathology and Genetics of Tumours of the Urinary System and Male Genital Organs. Lyon: IARC Press; 2004. World Health Organization Classification of Tumours. [Google Scholar]
- 18.Lin RK, Hsu HS, Chang JW, Chen CY, Chen JT, Wang YC. Alteration of DNA methyltransferases contributes to 5’CpG methylation and poor prognosis in lung cancer. Lung Cancer. 2007;55:205–213. doi: 10.1016/j.lungcan.2006.10.022. [DOI] [PubMed] [Google Scholar]
- 19.Yang J, Wei X, Wu Q, Xu Z, Gu D, Jin Y, Shen Y, Huang H, Fan H, Chen J. Clinical significance of the expression of DNA methyltransferase proteins in gastric cancer. Mol Med Report. 2011;4:1139–1143. doi: 10.3892/mmr.2011.578. [DOI] [PubMed] [Google Scholar]
- 20.Xing J, Stewart DJ, Gu J, Lu C, Spitz MR, Wu X. Expression of methylation-related genes is associated with overall survival in patients with non-small cell lung cancer. Br J Cancer. 2008;98:1716–1722. doi: 10.1038/sj.bjc.6604343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arai E, Kanai Y, Ushijima S, Fujimoto H, Mukai K, Hirohashi S. Regional DNA hypermethylation and DNA methyltransferase (DNMT) 1 protein overexpression in both renal tumors and corresponding nontumorous renal tissues. Int J Cancer. 2006;119:288–296. doi: 10.1002/ijc.21807. [DOI] [PubMed] [Google Scholar]
- 22.Wu CT, Wu CF, Lu CH, Lin CC, Chen WC, Lin PY, Chen MF. Expression and function role of DNA methyltransferase 1 in human bladder cancer. Cancer. 2011;117:5221–5233. doi: 10.1002/cncr.26150. [DOI] [PubMed] [Google Scholar]
- 23.Gravina GL, Ranieri G, Muzi P, Marampon F, Mancini A, Di Pasquale B, Di Clemente L, Dolo V, D’Alessandro AM, Festuccia C. Increased levels of DNA methyltransferases are associated with the tumorigenic capacity of prostate cancer cells. Oncol Rep. 2013;29:1189–1195. doi: 10.3892/or.2012.2192. [DOI] [PubMed] [Google Scholar]
- 24.Sawada M, Kanai Y, Arai E, Ushijima S, Ojima H, Hirohashi S. Increased expression of DNA methyltransferase 1 (DNMT1) protein in uterine cervix squamous cell carcinoma and its precursor lesion. Cancer Lett. 2007;251:211–219. doi: 10.1016/j.canlet.2006.11.023. [DOI] [PubMed] [Google Scholar]
- 25.Wu D, Xiong L, Wu S, Jiang M, Lian G, Wang M. TFPI-2 methylation predicts poor prognosis in non-small cell lung cancer. Lung Cancer. 2012;76:106–111. doi: 10.1016/j.lungcan.2011.09.005. [DOI] [PubMed] [Google Scholar]
- 26.Xu L, Li X, Chu ES, Zhao G, Go MY, Tao Q, Jin H, Zeng Z, Sung JJ, Yu J. Epigenetic inactivation of BCL6B, a novel functional tumor suppressor for gastric cancer, is associated with poor survival of gastric cancer. Gut. 2012;61:977–985. doi: 10.1136/gutjnl-2011-300411. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Q, Chen L, Helfand BT, Jang TL, Sharma V, Kozlowski J, Kuzel TM, Zhu LJ, Yang XJ, Javonovic B, Guo Y, Lonning S, Harper J, Teicher BA, Brendler C, Yu N, Catalona WJ, Lee C. TGF-b regulates DNA methyltransferase expression in prostate cancer, correlates with aggressive capabilities, and predicts disease recurrence. PLoS One. 2011;6:e25168. doi: 10.1371/journal.pone.0025168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Battagli C, Uzzo RG, Dulaimi E, Ibanez de Caceres I, Krassenstein R, Al-Saleem T, Greenberg RE, Cairns P. Promoter hypermethylation of tumor suppressor genes in urine from kidney cancer patients. Cancer Res. 2003;63:8695–8699. [PubMed] [Google Scholar]
- 29.Alleman WG, Tabios RL, Chandramouli GV, Aprelikova ON, Torres-Cabala C, Mendoza A, Rogers C, Sopko NA, Linehan WM, Vasselli JR. The in vitro and in vivo effects of re-expressing methylated von Hippel-Lindau tumor suppressor gene in clear cell renal carcinoma with 5-aza-2’deoxycytidine. Clin Cancer Res. 2004;10:7011–7021. doi: 10.1158/1078-0432.CCR-04-0516. [DOI] [PubMed] [Google Scholar]
- 30.Morris MR, Ricketts C, Gentle D, Abdulrahman M, Clarke N, Brown M, Kishida T, Yao M, Latif F, Maher ER. Identification of candidate tumor suppressor genes frequently methylated in renal cell carcinoma. Oncogene. 2010;29:2104–211. doi: 10.1038/onc.2009.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ting AH, Jair KW, Suzuki H, Yen RW, Baylin SB, Schuebel KE. Mammalian DNA methyltransferase 1: inspiration for new directions. Cell Cycle. 2004;3:1024–1026. [PubMed] [Google Scholar]
- 32.Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349:2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 33.Braconi C, Huang N, Patel T. MicroRNA-dependent regulation of DNA methyltransferase-1 and tumor suppressor gene expression by interleukin-6 in human malignant cholangiocytes. Hepatology. 2010;51:881–890. doi: 10.1002/hep.23381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fabbri M, Garzon R, Cimmino A, Liu Z, Zanesi N, Callegari E, Liu S, Alder H, Costinean S, Fernandez-Cymering C, Volinia S, Guler G, Morrison CD, Chan KK, Marcucci G, Calin GA, Huebner K, Croce CM. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci U S A. 2007;104:15805–15810. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Duursma AM, Kedde M, Schrier M, le Sage C, Agami R. miR-148 targets human DNMT3b protein coding region. RNA. 2008;14:872–877. doi: 10.1261/rna.972008. [DOI] [PMC free article] [PubMed] [Google Scholar]


