Abstract
The objective of the current study was to investigate the expression pattern and clinicopathological significance of Period1 (Per1), Period2 (Per2) and Period3 (Per3) in patients with non-small cell lung cancer (NSCLC). In 130 archived NSCLC tissues, the positive rate of Per1 (86/130, 66.2%), Per2 (77/130, 59.2%) and Per3 (82/130, 63.1%) were reduced in human lung cancer samples compared with adjacent normal lung tissues (Per1, 119/130, 91.5%; Per2, 115/130, 88.5%; Per3, 121/130, 93.1%), as measured by immunohistochemical staining. Loss of Per1 was correlated with poor differentiation (P < 0.001), tumor status (P=0.04), high p-TNM stage (P < 0.001) and lymph node metastasis (P=0.045). The similar tendencies were also found in the correlation of the expression of Per2 and Per3 with clinicopathological factors. In addition, a significant correlation was found between Per1 and Per2 (P < 0.001) in 130 cases of NSCLC. Similarly, a significant correlation was found between Per2 and Per3 (P=0.045). Patients with lower expression of Per1, Per2 and Per3 had a shorter survival time than those with higher expression. These results indicate that loss of Per may promote tumor progression in NSCLC, and may serve as a novel prognostic biomarker of NSCLC.
Keywords: Period1, Period2, Period3, non-small cell lung cancer, immunohistochemistry, prognosis
Introduction
Lung cancer, the most commonly diagnosed malignant tumor, is the leading cause of death among the malignant tumors throughout the world [1]. The incidence of lung cancer is still increasing in China due to smoking and air pollution, etc. Non-small cell lung cancer (NSCLC) is any type of epithelial lung cancer other than small cell lung cancer (SCLC). In comparison to SCLC, NSCLCs have better prognosis but are relatively insensitive to chemotherapy. Although surgical resection, chemotherapy and radiotherapy have been established, the long-term survival for lung cancer patients is still unsatisfying. Improvement in the understanding of molecular processes involved in pulmonary carcinogenesis has led to new treatment options such as targeted small molecules and vaccines. Therefore, better defining the pathogenesis of lung cancer, looking for useful biomarkers, and exploring novel therapeutic targets are demanding tasks.
Different types of living organisms are driven by the daily light-dark cycles of the earth. Such circadian rhythm is one of the basic characteristics of an organism’s life activities. Circadian rhythm is controlled by the circadian system, which is composed of serious circadian clock genes [2]. These clock genes regulate the much more biological effect beyond the circadian rhythm, such as the cell proliferation, apoptosis, immune response, hypoxia, angiogenesis, tumor genesis and tumor progression [3]. The common known core clock genes include CLOCK, BMAL1, Period1 (Per1), Period2 (Per2), Period3 (Per3), cryptochrome1 (Cry1), cryptochrome2 (Cry2), cascin kinase1 epsilon (CSNK1E) and timeless (TIM) [4]. Epidemiologic studies have shown that disruption of normal circadian rhythm may increase the risk of developing various types of cancer such as breast, prostate, colorectal, liver and endometrial cancers [5,6].
Of the entire known clock genes, the members of period (Per) subfamily have been shown to play a major role in cancer development. Overexpression of Per1 or Per2 in cancer cells inhibits their neoplastic growth and apoptosis [7]. Previous study have shown the aberrant expression and abnormal rhythms of Per1 were highly linked to the carcinogenesis and development of malignant tumors, such as prostate cancer, gastric cancer, colon cancer, leukocythemis, breast cancer and buccal squmaous cell carcinoma [5,8-12]. Recently, it has been reported that Per2 was mainly located in the nuclei of the Clara cells and the latter is critical for maintaining coherent circadian oscillations of the lung [13]. Gery et al. have reported that Per1 was down-regulated in NSCLC cell lines and inhibit the cell proliferation [14]. In addition, Couto et al. reported that Per3 was downregulated in NSCLC [15]. These reports indicate that the members of Per subfamily play a major role in the tumor progression in NSCLC.
In this study, we focused on the expression and clinicopathological significance of Per1, Per2 and Per3 in NSCLC.
Materials and methods
Patients and specimens
Ethical approval for this study was obtained from the local trials committee of the Liaoning Cancer Hospital and Institute. Primary tumor specimens were obtained from 130 patients (75 males and 55 females), who were diagnosed with lung squamous cell carcinoma (SCC) or adenocarcinoma and underwent complete resection in the Liaoning Cancer Hospital and Institute between 2006 and 2008. Large cell carcinoma, adenosquamous carcinoma, or other NSCLC subtypes were excluded in this study. None of the patients had received radiotherapy or chemotherapy before surgical resection, and all the patients were treated with routine chemotherapy after operation. The mean age of the patients was 61 years (range, 38-79 years). The histological diagnosis and grade of differentiation were evaluated using hematoxylin-eosin-stained sections according to the World Health Organization guidelines of classification. All 130 specimens were re-evaluated with respect to histological subtype, differentiation and tumor stage. SCC was identified in 62 of the cases, and adenocarcinoma was in 68 of the samples. Lymph node metastases were identified in 58 of the 130 patients. The p-TNM staging system of the International Union Against Cancer (7th edition) was used to classify specimens as stages I (n=57), II (n=31) and III (n=42). The patients were followed-up once every 3 months for the first 2 years, once every 6 months during the third to fifth years and annually for an additional 3 years or until postoperative patient death. All patients were contacted by phone to determine their health status, and the last follow-up date was January 1, 2014. The overall survival (OS), which was defined as the time from the operation to patient death or the last follow-up, was used as a measure of prognosis.
Immunohistochemistry
Surgically excised tumor specimens and adjacent normal tissues were fixed with 10% neutral formalin, embedded in paraffin, and cut into 4-m-thick sections. Immunostaining was performed using the avidin-biotin-peroxidase complex method (Ultrasensitive; MaiXin, Fuzhou, China). The sections were deparaffinized in xylene, rehydrated with graded alcohol, and then boiled in 0.01 M citrate buffer (pH 6.0) for 2 min with an autoclave. Hydrogen peroxide activity and the sections were incubated with normal goat serum to reduce non-specific binding. Tissue sections were incubated with Per1 rabbit polyclonal antibody (1:250 dilution, Abcam, Cambridge, MA, US), Per2 mouse monoclonal antibody (1:150 dilution, Abnova, Taipei, Taiwan) and Per3 mouse monoclonal antibody (1:100 dilution, Santa Cruz Biotechnology Inc. CA, USA). Rabbit and mouse immunoglobulin (at the same concentration of the antigen specific antibody) were used as isotype controls. Staining for primary antibodies was performed at room temperature for 2 h. A mixture of biotinylated goat anti-rabbit and goat anti-mouse serum IgG were used as secondary antibody. Following rinsing with PBS for three times, the sections were incubated with streptavidin-biotin conjugated with horseradish peroxidase, and the peroxidase reaction was developed with 3, 3’-diaminobenzidine tetrahydrochloride. Counterstaining with hematoxylin was performed and the sections were dehydrated in ethanol before mounting.
Two independent, blinded investigators examined all tumor slides. 100 cells were observed and scored per view at ×400 magnification. Both the proportion of nuclear positive cells and intensity were taken consideration due to the variation among different lesions. For each sample, the rate of positive cells was categorized as follows: 0, < 5%; 1, 5-25%; 2, 26-50%; 3, 51-75%; 4, > 75%. The intensity was graded as follows: 0, negative; 1, weak; 2, moderate; 3, strong. The proportion and intensity scores were then multiplied to obtain a final score. Scores of 0-4 were defined as “low expression”; scores of 5-12 as “high expression”. The criteria of evaluation for Per1, Per2 and Per3 are the same.
Statistical analysis
SPSS version 13.0 for windows was used for all analyses (SPSS, Chicago, IL, USA). The Pearson Chi-Square test was used to examine possible correlations between Per1, Per2, Per3 and clinicopathological factors. Survival curves were plotted using the Kaplan-Meier method and compared with the log-rank test. The association among Pers was assessed using Spearman’s correlation coefficient analysis. All p values were two-sided, and P < 0.05 was determined to be statistically significant.
Results
Per1, Per2 and Per3 expression pattern in normal lung tissues and NSCLC tissues
The characteristics of the 130 patients are summarized in Table 1. The expression of Per1, Per2 and Per3 were assessed by immunohistochemistry in 130 NSCLC samples with the matched adjacent noncancerous tissues. In the noncancerous tissues, Per1, Per2 and Per3 were predominantly located in the nucleus of the alveolar epithelial cells (Figure 1A-F), and normal bronchial epithelium (Figure 1G-I). Cells positive for Per1, Per2 or Per3 were more than 85%. However, the expression of Per1, Per2 and Per3 in cancer cells was frequently reduced compared with the adjacent normal bronchi epithelium. Per1, Per2 and Per3 localized predominantly in the nuclear, and can be observed in the cytoplasm (Figure 2A-R). As summarized in Table 2, out of 130 NSCLC samples, 33.8% (44/130) of them display a loss of Per1 expression and 66.2% of total samples (86/130) remain Per1 positive. However, in adjacent noncancerous tissues, Per1 was positively stained in 91.5% (119/130) of samples and only 8.5% (11/130) showed the loss of Per1 expression. The similar results were also found in Per2 and Per3 expression pattern in NSCLC compared with normal adjacent noncancerous tissues. The positive rate of Per2 and Per3 in NSCLC were 59.2% (77/130) and 63.1% (82/130), respectively. Collectively, NSCLC showed a significantly reduction in the expression of Per1, Per2 and Per3.
Table 1.
Clinical characteristics and Per1, Per2 and Per3 expression in 130 cancer patient samples
| Number of cases (%) | |
|---|---|
| Gender | |
| Male | 75 (57.7) |
| Female | 55 (42.3) |
| Age | |
| < 60 | 62 (47.7) |
| ≥ 61 | 68 (52.3) |
| Clinical stage | |
| I | 57 (43.8) |
| II | 31 (23.8) |
| III | 42 (32.2) |
| Lymph node metastasis | |
| N0 | 72 (55.4) |
| N1-3 | 58 (44.6) |
| Histological types | |
| well | 44 (33.8) |
| moderate | 64 (49.2) |
| poor | 22 (17.0) |
| Vital status | |
| Alive | 13 (10) |
| Death (All NSCLC-related) | 117 (90) |
| Expression of Per1 | |
| negative | 44 (33.8) |
| positive | 86 (66.2) |
| Expression of Per2 | |
| negative | 53 (40.8) |
| positive | 77 (59.2) |
| Expression of Per3 | |
| negative | 48 (36.9) |
| positive | 82 (63.1) |
NSCLC, non-small cell lung cancer.
Figure 1.
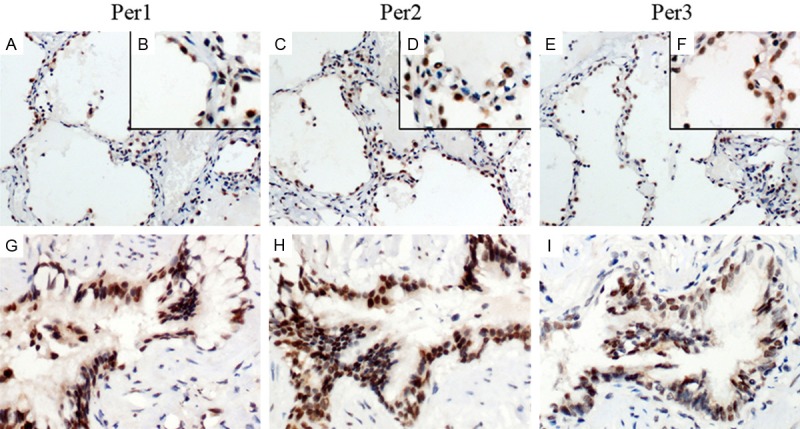
The expression pattern of Per1, Per2 and Per3 in normal lung tissues. In noncancerous tissues, Per1, Per2 and Per3 were predominantly located in the nucleus of the alveolar epithelial cells (A, C, E, magnification ×200; B, D, F, magnification ×400) and normal bronchial epithelium (G-I, magnification ×400). The amplification of local parts was placed in the upper right.
Figure 2.
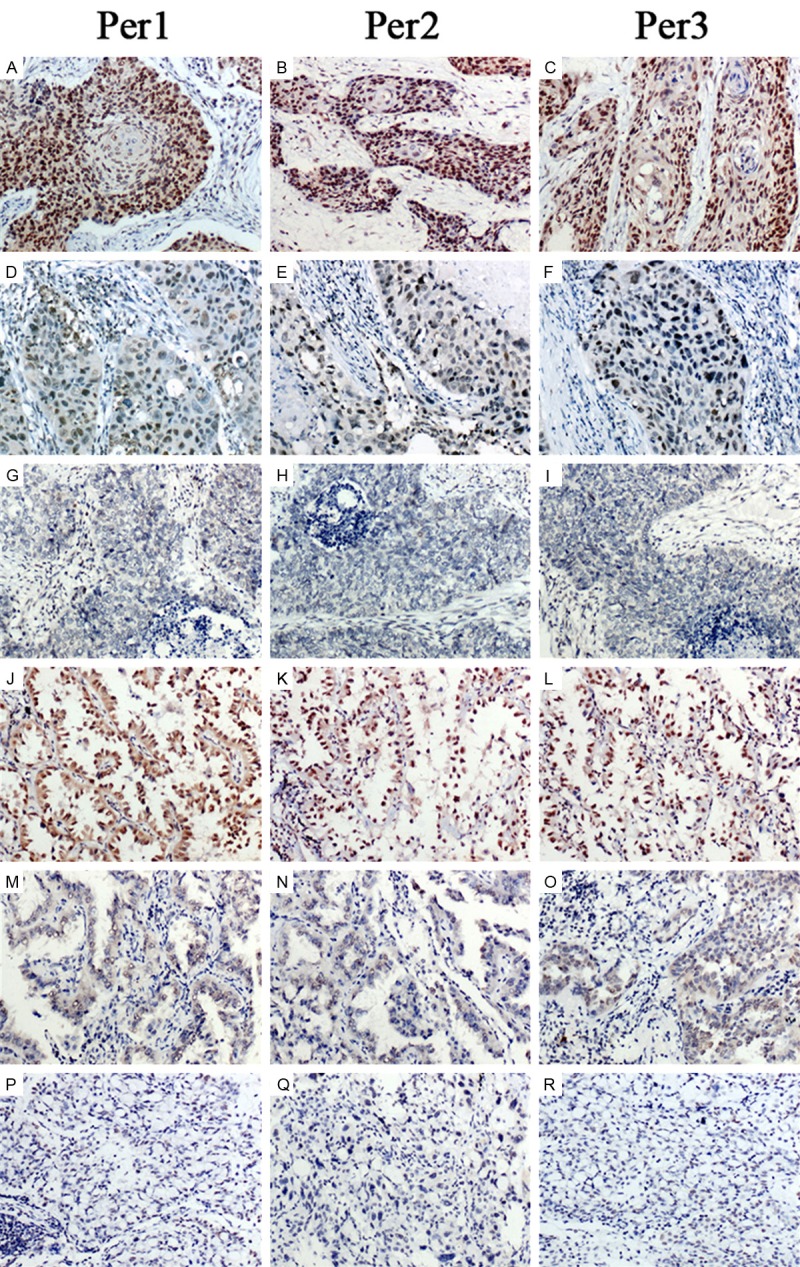
The expression pattern of Per1, Per2 and Per3 in NSCLC with different differentiation. All Pers were reduced accompany with the tumor progression. (A-I) squamous cell carcinoma (A-C represent well-differentiated SCC, D-F represent moderate-differentiated SCC, G-I represent poorly-differentiated SCC); (J-R) adenocarcinoma (J-L represent well-differentiated adenocarcinoma, M-O represent moderate-differentiated adenocarcinoma, P-R represent poorly-differentiated adenocarcinoma). Per1, Per2 and Per3 were reduced in moderate and poorly differentiated NSCLCs (D-I and M-R, respectively) compared with well-differentiated NSCLCs (A-C and J-L, respectively).
Table 2.
Per1, Per2 and Per3 nuclear expression in NSCLC and adjacent non-tumor tissues
| Per1 | Per2 | Per3 | ||||
|---|---|---|---|---|---|---|
|
|
|
|
||||
| low | high | low | high | low | high | |
| NSCLC | 44 (33.8%) | 86 (66.2%) | 53 (40.8%) | 77 (59.2%) | 48 (36.9%) | 82 (63.1%) |
| Adjacent non-tumor tissues | 11 (8.5%) | 119 (91.5%)* | 15 (11.5%) | 115 (88.5%)* | 9 (6.9%) | 121 (93.1%)* |
NSCLC, non-small cell lung cancer.
P < 0.05 indicates statistical significance.
Clinical significance of Pers expression in NSCLCs
The association of Per1, Per2 or Per3 expression and clinicopathological variables is described in Table 3. To further address the relationship between Per1, Per2 and Per3 with cancer, several clinicopathological factors in NSCLC were examined. As shown in Table 3, our statistical analysis showed that loss of Per1 was significantly correlated with tumor differentiation (P < 0.001), tumor status (T1+T2 vs. T3+T4, P=0.04), TNM stage (I+II vs. III, P < 0.001) and lymph node metastasis (P=0.045) in NSCLC. Loss of Per2 was significantly correlated with tumor differentiation (P=0.001), tumor status (T1+T2 vs. T3+T4, P=0.002), TNM stage (I+II vs. III, P < 0.001) and lymph node metastasis (P=0.003) in NSCLC. Loss of Per3 was also significantly correlated with tumor differentiation (P=0.005), tumor status (T1+T2 vs. T3+T4, p=0.044), TNM stage (I+II vs. III, P < 0.001) and lymph node metastasis (P=0.016) in NSCLC. Taken together, the decrease of Pers expression is linked to poor differentiation, tumor size, high TNM stage and lymph node metastasis. However, no significant associations were observed between Per1 level with patients’ age (P=0.995), gender (p=0.817) and histological type (P=0.268). Likewise, no significant correlations were observed between Per2 and Per3 with patients’ age, gender and histological type. As shown in Figure 2, high expression of Per1, Per2 and Per3 were correlated with well differentiation. In contrast, low expression of Per1, Per2 and Per3 were correlated with moderate or poor differentiation. In addition, a significant correlation was found between Per1 and Per2 (P < 0.001) in 130 cases of NSCLC. Similarly, a significant correlation was also found between Per2 and Per3 (P=0.045) (Table 4).
Table 3.
Summary of correlation of Per1, Per2 and Per3 expression with clinicopathological characteristics in NSCLC
| Characteristics | Per1 | Per2 | Per3 | ||||||
|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|||||||
| low | high | P-value | low | high | P-value | low | high | P-value | |
| Age (years) | |||||||||
| < 60 | 21 | 41 | 0.995 | 25 | 37 | 0.921 | 26 | 36 | 0.258 |
| ≥ 61 | 23 | 45 | 28 | 40 | 22 | 46 | |||
| Gender | |||||||||
| Male | 26 | 49 | 0.817 | 29 | 46 | 0.569 | 26 | 49 | 0.534 |
| Female | 18 | 37 | 24 | 31 | 22 | 33 | |||
| Histology | |||||||||
| Squamous cell carcinoma | 18 | 44 | 0.268 | 25 | 37 | 0.921 | 23 | 39 | 0.969 |
| Adenocarcinoma | 26 | 42 | 28 | 40 | 25 | 43 | |||
| Differentiation | |||||||||
| Well | 9 | 35 | < 0.001* | 13 | 31 | 0.001* | 10 | 34 | 0.005* |
| Moderate | 18 | 46 | 23 | 41 | 24 | 40 | |||
| poor | 17 | 5 | 17 | 5 | 14 | 8 | |||
| Tumor status | |||||||||
| T1+T2 | 28 | 69 | 0.04* | 32 | 65 | 0.002* | 31 | 66 | 0.044* |
| T3+T4 | 16 | 17 | 21 | 12 | 17 | 16 | |||
| TNM stage | |||||||||
| I+II | 20 | 68 | < 0.001* | 26 | 62 | < 0.001* | 22 | 66 | < 0.001* |
| III | 24 | 18 | 27 | 15 | 26 | 16 | |||
| Nodal status | |||||||||
| N0 | 19 | 53 | 0.045* | 21 | 51 | 0.003* | 20 | 52 | 0.016* |
| N1, N2, N3 | 25 | 33 | 32 | 26 | 28 | 30 | |||
NSCLC, non-small cell lung cancer.
P < 0.05 indicates statistical significance.
Table 4.
The correlation between Per2 and Per1/Per3 in NSCLC
| Per1 expression | Per3 expression | ||||||||
|---|---|---|---|---|---|---|---|---|---|
|
|
|
||||||||
| low | high | P-value | Correlation coefficient | low | high | P-value | Correlation coefficient | ||
| Per2 expression | low | 34 | 19 | < 0.001* | 0.531 | 25 | 28 | 0.045* | 0.176 |
| high | 10 | 67 | 23 | 54 | |||||
NSCLC, non-small cell lung cancer.
P < 0.05 indicates statistical significance.
Prognostic implications of Per1, Per2 and Per3 expression in NSCLC
Kaplan-Meier analysis was used to calculate the effects of Per1, Per2 and Per3 expression on survival. The NSCLC patients with low Per1, Per2 and Per3 expression had significantly shorter overall survival time than those with high Per1, Per2 and Per3 expression (Figure 3A-C, for all Pers, P < 0.001). The postoperative median OS of patients with high staining of Per1 was 59 months, while that of patients with low staining of Per1 was 28 months. The overall five-year accumulative survival rate was 50.8%. The 5-year cumulative survival rate of patients with Per1 negative expression was 7.7%, compared with 43.1% of patients with Per1 positive expression. The postoperative median OS of patients with high staining of Per2 was 63 months, while that of patients with low Per2 expression was 28 months. The postoperative median OS of patients with high staining of Per3 was 59 months, while that of patients with low Per3 expression was 33 months. The 5-year OS rate in patients exhibiting elevated Per2 were significantly higher than in those exhibiting reduced Per2 levels (43.1% vs. 7.7%). The 5-year OS rate in patients exhibiting elevated Per3 was significantly higher than in those exhibiting reduced Per3 levels (40% vs. 10.8%). When the survival of patients with higher expression of Per1/Per2/Per3 was compared with that of those with low Per1/Per2/Per3, Kaplan-Meier analysis revealed a significant difference on overall survival (P < 0.001, Figure 3D).
Figure 3.
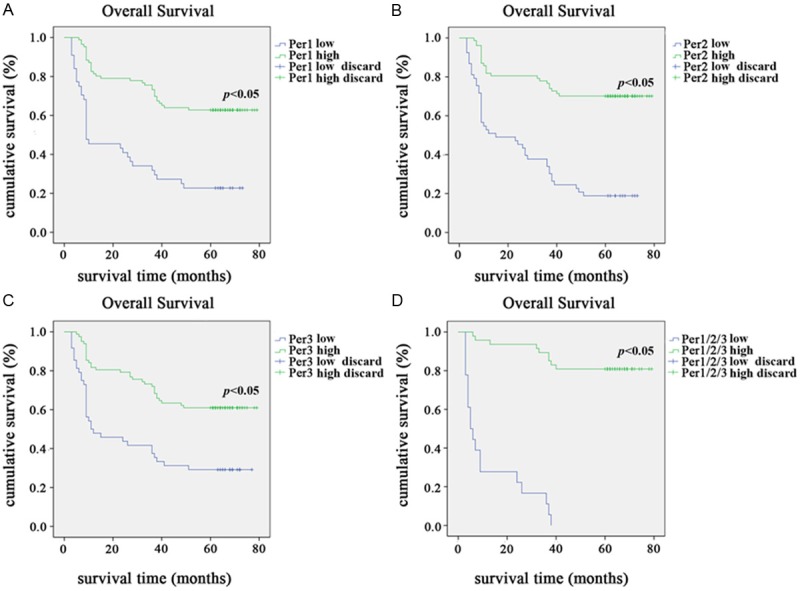
Kaplan-Meier survival curves for NSCLC patients with Per1, Per2 and Per3 negative expression (blue line) versus Per1, Per2 and Per3 positive expression (green line). A. The overall survival of patients (clinical stages I-III) with low/high Per1 expression; B. The overall survival of patients (clinical stages I-III) with low/high Per2 expression; C. The overall survival of patients (clinical stages I-III) with low/high Per3 expression; D. The patients with Per1/Per2/Per3 triple high expression revealed a significant difference in overall survival when compared with those patients with Per1/Per2/Per3 triple low expression.
Discussion
The results of this study show that the aberrant expression or loss of Per1, Per2 and Per3 is correlated with the poor differentiation, lymph node metastasis and high TNM stage in NSCLC. The patients with low expression of these genes have shorter overall survival time and lower 5-year cumulative survival rate than those patients with high expression of Per(s).
Per is a core clock gene which regulates the proliferation, secretion, metabolism of normal cells and circadian rhythm of living subjects. In mammals, there are three known Per family genes: Per1, Per2 and Per3. These clock genes also regulate the biological effect beyond the circadian rhythm, such as apoptosis, immune response, hypoxia and angiogenesis. Recently, it has been reported that Per1, Per2 and Per3 are involved in abnormal cell proliferation, carcinogenesis and development of malignant tumors, including breast cancer, prostate cancer, colorectal carcinoma and gliomas [5,8,16-18]. However, the expression and prognostic value of the Per1, Per2 and Per3 genes in NSCLC are largely unknown. Gibbs et al. found Per2 was located in the nuclei of the Clara cells and regulated the circadian oscillations of the lung [13]. They also demonstrated that the aberrant Per2 expression would induce some respiratory diseases. In addition, Gery et al. found the loss of Per1 inhibited the cell proliferation in NSCLC cell lines [14]. Patricia et al. also found the polymorphism in Per3 might be a risk factor in the occurrence and development of NSCLC [15].
Aiming to investigate the role of Per in the carcinogenesis in NSCLC, we performed immunohistochemistry staining to detect the expression of Per1, Per2 and Per3 in 130 cases of NSCLC and adjacent non-tumorous tissues. We studied the relations between the expression levels of these genes and clinicopathological features. In addition, we also performed Kaplan-Meier analysis to investigate the correlation of these genes with prognosis.
We noticed that several previous studies have explored the expression and clinical significance of Per1, Per2 and Per3 in other types of cancer. In colon cancer, breast cancer and oral SCC, the expression of Per1 was lower than that in normal adjacent tissues [18-20]. Similar tendency is also found in Per2 expression pattern in breast cancer [18]. Additionally, Per1 was correlated with stages of disease, depth of invasion and the presence of lymph node metastasis [20]. These results indicate loss of Per1 expression was associated with the progression and malignancy of tumors [20].
Our results are in accord with those results described above. We also found that low expression of Per1, Per2 and Per3 were more frequently observed in lung cancers compared with those in normal adjacent noncancerous tissues. The loss of Per1, Per2 and Per3 were associated with poor differentiation, high TNM stage and lymph node metastasis in NSCLC. Moreover, the expression of Per1 was significantly correlated with the expression of Per2, and the latter was also correlated with the expression of Per3. In addition, Kaplan-Meier analysis showed that, in general, patients with high Per1, Per2 and Per3 expression experienced a better survival outcome, while patients with low Per1, Per2 and Per3 expression had a poorer survival outcome. Our results showed the first evidence that Per2 was correlated with both Per1 and Per3. Thus, Per1, Per2, Per3 might play a potential role in suppressing the progression and metastasis of NSCLC.
Based on our results, Per seems to be a cancer suppressor in the carcinogenesis in NSCLC. It has been reported that Per functions as an pro-apoptotic factor in cancer cells [21], and this well explained that the loss of Per may cause the resistance to the apoptosis in the poor differentiated NSCLC. However, the detailed mechanism of the down-regulation of Per is still a problem to be explored. As well known, CpG methylation of promoter sequences, particularly in tumor suppressor or related genes, can inactivate promoter functions and lead to silencing of these genes. Methylation is commonly convicted as a critical causal event in silencing the cancer suppressor gene [22]. Therefore, we speculate that the inhibition of Per is caused by the methylation in the promoters of Per genes. Chen et al. reported that the aberrant expression of Per in breast cancer is caused by methylation of the PER1 or PER2 promoter instead of genetic mutations. However, Remco et al. identified the miR-192/194 cluster as a potent inhibitor of the entire Period gene family using a forward genetic screen, which indicated a new mechanism for the down-regulation of Per at the post-transcriptional level [23]. Recently, Couto et al. reported five SNPs in the Per3 in NSCLC patients [15]. Based on the reports above, we suspect that the cause of the inhibition of Per genes in caners is complicated, and methylation may be not the only reason. The mechanisms leading to the inhibition of Per genes is still largely unknown. Thus, the potential role of Per in NSCLC development and their underlying mechanisms should be investigated further.
Acknowledgements
This study was supported by grant from Key Programs of Science and Technology of Liaoning Province (2002225001-5&2010225032).
Disclosure of conflict of interest
None.
References
- 1.Huang B, Carloss H, Wyatt SW, Riley E. Hormone replacement therapy and survival in lung cancer in postmenopausal women in a rural population. Cancer. 2009;115:4167–4175. doi: 10.1002/cncr.24475. [DOI] [PubMed] [Google Scholar]
- 2.Hara Y, Onishi Y, Oishi K, Miyazaki K, Fukamizu A, Ishida N. Molecular characterization of Mybbp1a as a co-repressor on the Period2 promoter. Nucleic Acids Res. 2009;37:1115–1126. doi: 10.1093/nar/gkn1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu Y, Wang L, Lin XY, Wang J, Yu JH, Miao Y, Wang EH. The transcription factor DEC1 (BHLHE40/STRA13/SHARP-2) is negatively associated with TNM stage in non-small-cell lung cancer and inhibits the proliferation through cyclin D1 in A549 and BE1 cells. Tumour Biol. 2013;34:1641–1650. doi: 10.1007/s13277-013-0697-z. [DOI] [PubMed] [Google Scholar]
- 4.Guillaumond F, Dardente H, Giguere V, Cermakian N. Differential control of Bmal1 circadian transcription by REV-ERB and ROR nuclear receptors. J Biol Rhythms. 2005;20:391–403. doi: 10.1177/0748730405277232. [DOI] [PubMed] [Google Scholar]
- 5.Cao Q, Gery S, Dashti A, Yin D, Zhou Y, Gu J, Koeffler HP. A role for the clock gene per1 in prostate cancer. Cancer Res. 2009;69:7619–7625. doi: 10.1158/0008-5472.CAN-08-4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao B, Lu J, Yin J, Liu H, Guo X, Yang Y, Ge N, Zhu Y, Zhang H, Xing J. A functional polymorphism in PER3 gene is associated with prognosis in hepatocellular carcinoma. Liver Int. 2012;32:1451–1459. doi: 10.1111/j.1478-3231.2012.02849.x. [DOI] [PubMed] [Google Scholar]
- 7.Fu L, Lee CC. The circadian clock: pacemaker and tumour suppressor. Nat Rev Cancer. 2003;3:350–361. doi: 10.1038/nrc1072. [DOI] [PubMed] [Google Scholar]
- 8.Mostafaie N, Kallay E, Sauerzapf E, Bonner E, Kriwanek S, Cross HS, Huber KR, Krugluger W. Correlated downregulation of estrogen receptor beta and the circadian clock gene Per1 in human colorectal cancer. Mol Carcinog. 2009;48:642–647. doi: 10.1002/mc.20510. [DOI] [PubMed] [Google Scholar]
- 9.Yeh KT, Yang MY, Liu TC, Chen JC, Chan WL, Lin SF, Chang JG. Abnormal expression of period 1 (PER1) in endometrial carcinoma. J Pathol. 2005;206:111–120. doi: 10.1002/path.1756. [DOI] [PubMed] [Google Scholar]
- 10.Hsu CM, Lin SF, Lu CT, Lin PM, Yang MY. Altered expression of circadian clock genes in head and neck squamous cell carcinoma. Tumour Biol. 2012;33:149–155. doi: 10.1007/s13277-011-0258-2. [DOI] [PubMed] [Google Scholar]
- 11.Yang MY, Yang WC, Lin PM, Hsu JF, Hsiao HH, Liu YC, Tsai HJ, Chang CS, Lin SF. Altered expression of circadian clock genes in human chronic myeloid leukemia. J Biol Rhythms. 2011;26:136–148. doi: 10.1177/0748730410395527. [DOI] [PubMed] [Google Scholar]
- 12.Yang X, Wood PA, Ansell CM, Quiton DF, Oh EY, Du-Quiton J, Hrushesky WJ. The circadian clock gene Per1 suppresses cancer cell proliferation and tumor growth at specific times of day. Chronobiol Int. 2009;26:1323–1339. doi: 10.3109/07420520903431301. [DOI] [PubMed] [Google Scholar]
- 13.Gibbs JE, Beesley S, Plumb J, Singh D, Farrow S, Ray DW, Loudon AS. Circadian timing in the lung; a specific role for bronchiolar epithelial cells. Endocrinology. 2009;150:268–276. doi: 10.1210/en.2008-0638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gery S, Komatsu N, Kawamata N, Miller CW, Desmond J, Virk RK, Marchevsky A, McKenna R, Taguchi H, Koeffler HP. Epigenetic silencing of the candidate tumor suppressor gene Per1 in non-small cell lung cancer. Clin Cancer Res. 2007;13:1399–1404. doi: 10.1158/1078-0432.CCR-06-1730. [DOI] [PubMed] [Google Scholar]
- 15.Couto P, Miranda D, Vieira R, Vilhena A, De Marco L, Bastos-Rodrigues L. Association between CLOCK, PER3 and CCRN4L with nonsmall cell lung cancer in Brazilian patients. Mol Med Rep. 2014;10:435–440. doi: 10.3892/mmr.2014.2224. [DOI] [PubMed] [Google Scholar]
- 16.Chen ST, Choo KB, Hou MF, Yeh KT, Kuo SJ, Chang JG. Deregulated expression of the PER1, PER2 and PER3 genes in breast cancers. Carcinogenesis. 2005;26:1241–1246. doi: 10.1093/carcin/bgi075. [DOI] [PubMed] [Google Scholar]
- 17.Wang Y, Hua L, Lu C, Chen Z. Expression of circadian clock gene human Period2 (hPer2) in human colorectal carcinoma. World J Surg Oncol. 2011;9:166. doi: 10.1186/1477-7819-9-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Winter SL, Bosnoyan-Collins L, Pinnaduwage D, Andrulis IL. Expression of the circadian clock genes Per1 and Per2 in sporadic and familial breast tumors. Neoplasia. 2007;9:797–800. doi: 10.1593/neo.07595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oshima T, Takenoshita S, Akaike M, Kunisaki C, Fujii S, Nozaki A, Numata K, Shiozawa M, Rino Y, Tanaka K, Masuda M, Imada T. Expression of circadian genes correlates with liver metastasis and outcomes in colorectal cancer. Oncol Rep. 2011;25:1439–1446. doi: 10.3892/or.2011.1207. [DOI] [PubMed] [Google Scholar]
- 20.Chen R, Yang K, Zhao NB, Zhao D, Chen D, Zhao CR, Tang H. Abnormal expression of PER1 circadian-clock gene in oral squamous cell carcinoma. Onco Targets Ther. 2012;5:403–407. doi: 10.2147/OTT.S38508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gery S, Komatsu N, Baldjyan L, Yu A, Koo D, Koeffler HP. The circadian gene per1 plays an important role in cell growth and DNA damage control in human cancer cells. Mol Cell. 2006;22:375–382. doi: 10.1016/j.molcel.2006.03.038. [DOI] [PubMed] [Google Scholar]
- 22.Clark SJ, Melki J. DNA methylation and gene silencing in cancer: which is the guilty party? Oncogene. 2002;21:5380–5387. doi: 10.1038/sj.onc.1205598. [DOI] [PubMed] [Google Scholar]
- 23.Nagel R, Clijsters L, Agami R. The miRNA-192/194 cluster regulates the Period gene family and the circadian clock. FEBS J. 2009;276:5447–5455. doi: 10.1111/j.1742-4658.2009.07229.x. [DOI] [PubMed] [Google Scholar]


