Abstract
Parasitic protozoa, such as Leishmania species, are thought to express a number of surface and secreted nucleoside triphosphate diphosphohydrolases (NTPDases) which hydrolyze a broad range of nucleoside tri- and diphosphates. However, the functional significance of NTPDases in parasite virulence is poorly defined. The Leishmania major genome was found to contain two putative NTPDases, termed LmNTPDase1 and 2, with predicted NTPDase catalytic domains and either an N-terminal signal sequence and/or transmembrane domain, respectively. Expression of both proteins as C-terminal GFP fusion proteins revealed that LmNTPDase1 was exclusively targeted to the Golgi apparatus, while LmNTPDase2 was predominantly secreted. An L. major LmNTPDase1 null mutant displayed increased sensitivity to serum complement lysis and exhibited a lag in lesion development when infections in susceptible BALB/c mice were initiated with promastigotes, but not with the obligate intracellular amastigote stage. This phenotype is characteristic of L. major strains lacking lipophosphoglycan (LPG), the major surface glycoconjugate of promastigote stages. Biochemical studies showed that the L. major NTPDase1 null mutant synthesized normal levels of LPG that was structurally identical to wild type LPG, with the exception of having shorter phosphoglycan chains. These data suggest that the Golgi-localized NTPase1 is involved in regulating the normal sugar-nucleotide dependent elongation of LPG and assembly of protective surface glycocalyx. In contrast, deletion of the gene encoding LmNTPDase2 had no measurable impact on parasite virulence in BALB/c mice. These data suggest that the Leishmania major NTPDase enzymes have potentially important roles in the insect stage, but only play a transient or non-major role in pathogenesis in the mammalian host.
Author Summary
Nucleoside triphosphate diphosphohydrolases (NTPDases) are a family of enzymes expressed in many eukaryotes, ranging from single-celled parasites to mammals. In mammals, NTPDases can have an immunomodulatory role, while in pathogenic protists cell-surface and secreted NTPDases are thought to be important virulence factors, although this has never been explicitly tested. In this study we have investigated the function of two NTPDases, termed LmNTPDase1 and LmNTPDase2, in Leishmania major parasites. We show that LmNTPDase 1 and LmNTPDase 2 are differentially targeted to the Golgi apparatus and secreted, respectively. A Leishmania major mutant lacking the Golgi LmNTPDase1 exhibited a delayed capacity to induce lesions in susceptible mice when promastigote (insect) stages were used to initiate infection, but not when amastigote (mammalian-infective) stages were used. Loss of promastigote infectivity in the LmNTPDase1 null mutant was associated with the synthesis and surface expression of lipophosphoglycan (LPG), with shorter glycan chains and increased sensitivity to complement-mediated lysis. In contrast, a null mutant lacking the secreted LmNTPDase2 did not exhibit any difference in virulence. Our results suggest that Leishmania major NTPDases have specific roles in regulating Golgi glycosylation pathways, and nucleoside salvage pathways in the insect stages, but do not appear to be required for virulence of the mammalian-infective stages.
Introduction
Leishmania parasites cause a spectrum of diseases in humans, ranging from localized cutaneous lesions to disseminated mucocutaneous and lethal visceral infections. It is estimated that 1.5 to 2 million new cases of leishmaniasis occur annually and that more than 350 million people are at risk worldwide. Current first-line drug treatments are suboptimal due to high toxicity, cost, requirement for hospitalization and/or the emergence of drug-resistant strains, highlighting the need for the development of more effective therapeutics [1]. Leishmania parasites develop as extracellular promastigote stages in the digestive tract of the sandfly vector [2]. Following injection into the mammalian host during a sandfly bloodmeal, promastigotes are phagocytosed by a range of host cells (neutrophils, dendritic cells and macrophages) before differentiating to obligate intracellular amastigote stages that primarily proliferate within the phagolysosome compartment of macrophages. A number of surface molecules, including an abundant lipophosphoglycan (LPG) and several GPI-anchored glycoproteins, have been shown to be important for promastigote survival during these initial stages of infection [3]. In particular, LPG is thought to form a continuous surface glycocalyx that protects the promastigote stages of most Leishmania species from complement-mediated lysis and macrophage-induced oxidative stress during phagocytosis [3]–[5]. However, expression of LPG is down-regulated in amastigote stages and neither LPG nor GPI-anchored proteins are required for the long term growth and survival of this stage in macrophages. The potential role of other promastigote and amastigote secreted and surface proteins in the initiation and establishment of infection is less well defined.
A number of protozoan parasites have been shown to express nucleoside triphosphate diphosphohydrolase activities on their cell surface or in the extracellular milieu [6]–[9], and it has been suggested that hydrolysis of nucleotides may play a role in parasite pathogenesis [10]–[12]. Nucleoside triphosphate diphosphohydrolases (NTPDases, CD39_GDA1 protein superfamily) are a family of enzymes defined by the presence of five apyrase conserved regions (ACRs) and the ability to hydrolyze a wide range of nucleoside tri- and di-phosphates [13]. In mammals, surface-expressed NTPDases function in inflammation and immunity, vascular hemostasis and purine salvage [14], while in the intracellular bacterial pathogen, Legionella pneumophila, a secreted NTPDase is required for full virulence in a mouse model of disease [15], [16]. In Leishmania species, enzyme activity consistent with the presence of one or more surface-located NTPDases has been observed in both L. amazonensis and L. tropica, two species responsible for cutaneous leishmaniasis [17]–[19]. A number of lines of indirect evidence suggest that this surface NTPDase activity is important for virulence in the mammalian host. Specifically, surface NTPDase activity is elevated in virulent Leishmania strains and in the intracellular amastigote form of the parasite [17]–[19]; inhibition of surface NTPDase activity with chromium (III) adenosine 5′-triphosphate complex, reduced promastigote attachment and entry into mouse macrophages [20]; treatment of parasites with an antibody to the human NTPDase CD39 also reduced the interaction of Leishmania with mouse macrophages [19]; finally, polyclonal antibodies raised against synthetic peptides derived from the amino acid sequences of a putative L. braziliensis NTPDase caused significant cytotoxicity in cultured L. braziliensis promastigotes [21]. While these studies suggest roles for NTPDases in parasite nutrition, surface/secreted NTPDases could also contribute to pathogenesis by inducing host cell purinergic receptors. Purinergic receptors are upregulated in macrophages infected with L. amazonensis and these receptors display increased sensitivity to activation by nucleoside triphosphates (NTPs). As changes in the levels of extracellular NTPs and NDPs have been shown to alter purinergic receptor activity and the immune response [22], [23], it has been speculated that hydrolysis of host nucleotides by parasite ecto-NTPDases may restrict the immune response and facilitate parasite proliferation.
While these studies suggest NTPDases may function in Leishmania virulence and/or be essential for normal growth and development, they have relied heavily on techniques such as anti-NTPDase antibodies and/or chemical inhibition of enzyme activity to investigate the role of NTPDases in host-parasite interaction. Definitive genetic evidence of a relationship between a parasite NTPDase and parasite virulence is lacking. In this study, we show that L. major encodes two NTPDases, termed LmNTPDase1 and LmNTPDase2 (abbreviated to NTPD1 and NTPD2), and we generate null mutants in order to investigate their function during infection of mammalian cells. Our findings suggest that NTPD1 is primarily located to the Golgi apparatus, and plays an important role in regulating both the maturation of surface LPG and the capacity of L. major promastigotes to initially establish lesions. In contrast, NTPD2 was secreted, and was not required for lesion development, suggesting that its primary role is in the sandfly vector.
Methods
Ethics statement
Use of mice in this study was approved by the Institutional Animal Care and Use Committee of the University of Melbourne (ethics number 1212647.1). All animal experiments were performed in accordance with the Australian National Health Medical Research council guidelines (Australian code of practice for the care and use of animals for scientific purposes, 8th Edition, 2013, ISBN: 1864965975).
Bioinformatic analysis of putative NTPDases
Putative NTPDases were identified by BLAST [24] searching of the available Leishmania genomes, with subsequent manual identification of the conserved ACRs [25], [26]. Protein sequence alignments were performed using ClustalW [27], [28]. SMART [29], [30] was used to identify motifs within the protein sequences.
Parasite strains and culture conditions
L. major substrain MHOM/SU/73/5-ASKH was used to create all mutant and transfected lines. Parasites were routinely cultured as axenic promastigotes in Medium-199 (M199, Gibco, Invitrogen, Australia) supplemented with 10% heat-inactivated foetal bovine serum (FBS, Invitrogen) at 27°C or, prior to mouse infection and LPG purification, in SDM-79 medium supplemented with 10% FBS. G418 (Invitrogen, 100 µg mL−1) or nourseothricin (Werner BioAgents, Germany, 100 µg mL−1) was used as appropriate to maintain selection pressure on parasites transfected with pXGFP+-derived plasmids or pIR1SAT-derived and pXGSAT-derived plasmids, while puromycin (Invitrogen, 20 µg mL−1), hygromycin (Boehringer Mannheim, 100 µg mL−1) and bleocin (Calbiochem, 10 µg mL−1) were used to select transformants during mutagenesis. Lesion amastigotes were isolated by disrupting murine lesions (diameter 5–10 mm) by passage through a 70 µm plastic sieve, followed by passage through a 27 G needle to lyse macrophages and release parasites [31]. Cell debris was removed by slow speed centrifugation (50×g, 10 min, 4°C) and the supernatant centrifuged (2000×g, 10 min, 4°C) to collect amastigotes. Amastigotes were washed once in PBS and counted using a haemocytometer prior to use in mouse infections.
Genetic manipulation of L. major
Primer sequences used in genetic manipulation are detailed in supporting information (S1 Table). L. major NTPDase null mutants were created via sequential homologous gene replacement in a manner similar to that previously described [32], [33]. All L. major PCR products described below were obtained by amplification from genomic DNA. To delete ntpd1, an 854 bp 5′ untranslated region (UTR) containing a 5′ Asp718 site and a 3′ XhoI site was amplified, and a 805 bp 3′ UTR region containing a 5′ BamHI and a 3′ SacI site was amplified. These products were then sequentially cloned into the pBluescript II SK vector (Stratagene, CA, USA). Puromycin or hygromycin resistance cassettes were then excised from pXG-PAC and pXG-HYG [34] respectively and cloned into the XhoI/BamHI sites. To functionally delete ntpd2 a 688 bp fragment of the 5′ gene end was amplified with a 5′HindIII site and a 3′ BamHI/EcoRI/linker region, and an 1156 bp 3′ UTR region containing a 5′ BamHI/EcoRI/linker region and 3′ NotI site was amplified. An overlap PCR was then performed using these PCR products as template and the resultant product cloned into the HindIII/NotI sites of the pBluescript II SK vector (Stratagene, CA, USA). Puromycin and bleocin resistance cassettes were excised from pXG-PAC and pXG-PHLEO [34] respectively using BamHI and EcoRI, and cloned into the engineered BamHI/EcoRI sites. Deletion mutant constructs were verified by restriction digest profiles and DNA sequencing. Targeting constructs were then excised by KpnI/SapI (ntpd1) or HindIII/NotI (ntpd2) digest, gel purified and 5 µg of each sequentially electroporated into L. major as described previously [35]. Clonal transfectants resistant to both selection drugs were chosen and deletion of the target gene and integration of resistance cassettes confirmed via triplicate PCR. To generate the pIR1SAT-ntpd1 construct used in chromosomal complementation, full-length ntpd1 was excised from pXG-LmNTPDase1-GFP using BamHI and cloned into the BglII site of the pIR1SAT vector [36], [37]. SwaI digest was used to excise 5 µg of targeting DNA for electroporation into L. major Δntpd1. Clonal transformants were selected on basis of resistance to nourseothricin and incorporation into the ssu locus confirmed by PCR. To create the LmNTPDase-GFP fusion proteins, full length ntpd genes were individually cloned into pXG-GFP+ [38]. To express the LPG1-mCherry fusion protein, mCherry from pEGFP-mCherry-N1 [39] was amplified with a 5′SmaI/BglII site and 3′BamHI site and cloned into the SmaI/BamHI sites of pXGSAT, generating pXGSAT-mCherry. lpg1 [40] was amplified and then cloned into SmaI/BglII of pXGSAT-mCherry, creating pXG-LPG1-mCherry. The resulting constructs were confirmed via DNA sequencing and electroporated into wild type L. major as previously described [35].
Subcellular localization of LmNTPDase-GFP fusion proteins using immunoblotting and microscopy
Promastigotes were incubated in serum-free media for 24 hours before harvesting by high speed centrifugation (16000×g, 5 min). Supernatants were filtered through a 0.45 µM filter to remove intact parasites before supernatant proteins were precipitated with 10% trichloroacetic acid. The pellet and supernatant fractions were analyzed by standard SDS-PAGE and immunoblotting techniques, with LmNTPDase-GFP fusion proteins detected using anti-GFP antibody (clones 7.1 and 13.1, Roche, Germany) at 1∶1000 dilution. For microscopy studies live cells were immobilized on poly-L-lysine coated coverslips. Cells were visualized and images acquired using a Deltavision Elite fluorescent microscope and SoftWorx software.
Purification and biochemical analysis of LPG
Stationary phase promastigotes grown in SDM-79 supplemented with 10% FBS were harvested by centrifugation and LPG extracted from de-lipidated cells and purified using octyl-Sepharose chromatography, as described previously [41], [42]. The molecular weight of LPG was assessed via SDS-PAGE and silver staining using standard techniques. LPG was depolymerised with 40 mM trifluoroacetic acid (8 min, 100°C) and dephosphorylated with calf intestinal alkaline phosphatase. The repeat units were desalted by passage over a small column of AG 50-X12 (H+) over AG 4-X4 (OH-) (200 µL of each resin, Biorad) and chromatographed by high performance anion-exchange chromatography (HPAEC). The HPAEC system was equipped with a Dionex GP-50 gradient pump, a Carbo Pac PA-1 column (4×250 mm), with a PA-1 guard column and an ED50 integrated pulsed amperometric detector. The system was controlled and data analyzed by Chromeleon version 6.50 software (DIONEX). The eluents used in the system were 75 mM NaOH (E1) and 75 mM NaOH in 250 mM NaOAc (E2). Elution was performed by the following gradient: T0 = 0% (v/v) E2; T5 = 0% (v/v) E2; T40 = 100% (v/v) E2, T60 = 100% (v/v) E2, at a flow rate of 0.6 mL/minute. The phosphatidylinositol moiety of purified LPG was released by nitrous acid deamination (0.25 M sodium nitrite in 0.05 M sodium acetate buffer, pH 4.0; incubated at 40°C for 2.5 h), recovered by partitioning into water-saturated 1-butanol and analyzed using liquid chromatography mass spectrometry (LC/MS).
Peanut agglutinin assay
Washed stationary phase parasites (107 mL−1) were incubated with varying concentrations of peanut agglutinin (PNA) in PBS with 1% bovine serum albumin for 30 minutes at room temperature, and non-agglutinated parasites were counted using a haemocytometer (adapted from [43]).
Serum sensitivity assay
Serum sensitivity assays were performed in a similar manner to those previously described [5]. Stationary phase promastigotes were washed and resuspended in PBS (107 cells in 500 µL PBS with 1 µg mL−1 propidium iodide) and incubated with varying concentrations of human sera for 30 minutes. Fluorescence (indicating cell lysis) was then measured by flow cytometry.
Mouse model of cutaneous leishmaniasis
Virulence in mice was assessed using the tail base model of cutaneous leishmaniasis, as described previously [31]. Female BALB/c mice (6–8 week old, age-matched) were injected subcutaneously at the tail base. Lesion size was assessed weekly and scored 0–4, as described previously [44]. All parasite cell lines were passaged previously in mice to ensure no loss of virulence unrelated to the known genetic mutations. Parasites were re-isolated from mice as described in the “Parasite strains and culture conditions” section.
Statistical analysis
Unpaired, two-tailed t-tests were performed using Prism GraphPad software (version 6) and a P value less than 0.05 was considered significant. The exception was when more than two parasite strains were compared, in which case a two-way ANOVA, also using Prism GraphPad software, was performed to simultaneously compare the three different groups. A P value less than 0.05 was considered significant when comparing the differences between the three groups.
Results
L. major encodes two putative NTPDases that are conserved amongst Leishmania species
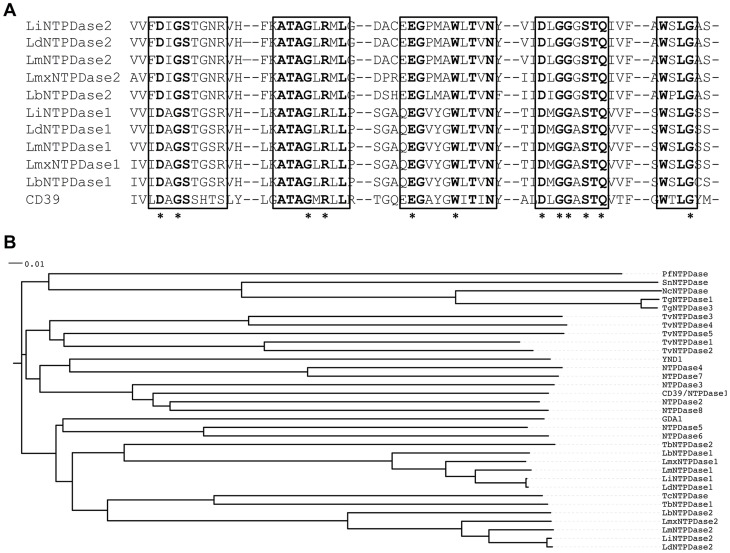
The L. major genome contains two putative NTPDase genes (LmjF15.0030 and LmjF10.0170), which are predicted to encode proteins with five ACR domains, the defining feature of all prokaryotic and eukaryotic NTPDase [45]. These genes are conserved amongst all sequenced Leishmania species, with homologues present in L. infantum, L. braziliensis, L. donovani and L. mexicana [46]. Importantly, a number of residues necessary for enzymatic activity of either CD39 or NTPDase3, the two best characterized mammalian NTPDases [47] are absolutely conserved within the Leishmania proteins (Fig. 1A). Using the nomenclature that we previously proposed for the parasite NTPDases [25], we refer to LmjF15.0030 as LmNTPDase1, and Lmj10.0170 as LmNTPDase2 (abbreviated to NTPD1 and NTPD2 in this study for succinctness). Homologues for NTPD1 and NTPD2 are present in T. brucei, but only NTPD2 exists in T. cruzi (Fig. 1B). Phylogenetic comparison with NTPDases found in other protozoa, mammals and yeast indicates that the trypanosomatid NTPDases are most closely related to mammalian NTPDase5 and NTPDase6, which are usually located intracellularly but can undergo secretion, and to the Golgi-located yeast NTPDase GDA1. Interestingly, the trypanosomatid NTPDases seem evolutionarily distinct from the NTPDases found in a range of apicomplexan parasites and Trichomonas protozoa (Fig. 1B), perhaps indicating divergent functions.
Figure 1. A. Alignment of regions of the putative Leishmania NTPDases with human CD39 (NTPDase1).
The conserved ACRs are aligned and boxed, with absolutely conserved residues shown in bold. Residues known to be necessary for enzyme function in mammalian NTPDases are starred, revealing all are present in the putative Leishmania NTPDases. Alignment was performed using ClustalW [27], [28]. B. Phylogenetic tree of protozoan, yeast and mammalian NTPDases. The tree was constructed from a ClustalW alignment of NTPDase amino acid sequences and viewed and edited using the Interactive Tree of Life web tool [63], [64]. Sequence accession numbers used for Fig. 1A and 1B are given in supplementary S2 Table.
NTPD1 localizes to the Golgi apparatus whereas NTPD2 is secreted from the parasite into the culture supernatant
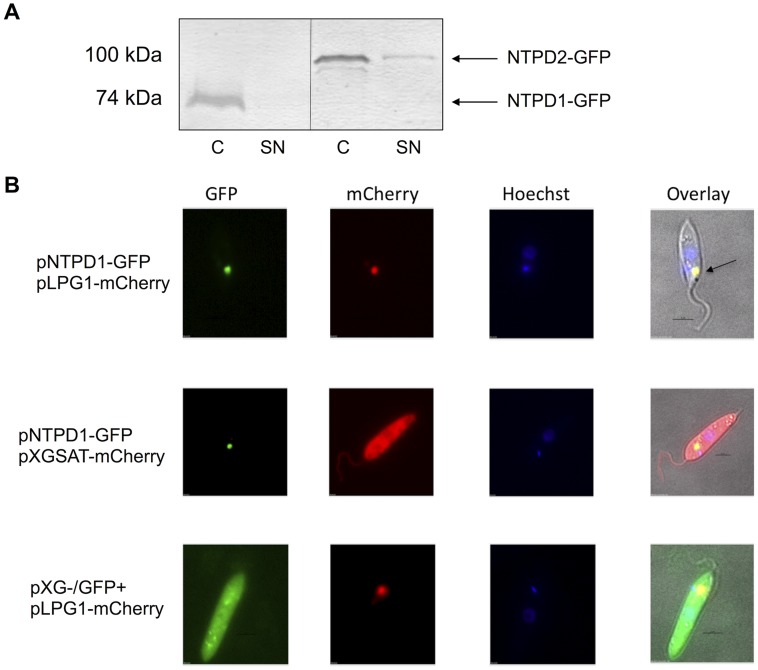
ntpd1 encodes for a protein (432 amino acids) with a putative N-terminal transmembrane domain (residues 17–36), while ntpd2 encodes for a longer protein (685 amino acids) with an N-terminal signal sequence (residues 1–20). To establish whether the two L. major NTPDases are secreted or targeted to the cell surface/intracellular compartment, wild type parasites were transfected with plasmids encoding NTPD1 and NTPD2 as fusion proteins containing C-terminal GFP. Western blot analysis of parasite cell pellets and culture supernatant showed that full-length proteins were expressed in each parasite line (Fig. 2A). Interestingly, while the NTPD1-GFP fusion protein was exclusively associated with the cell pellet, NTPD2-GFP fusion protein was secreted (Fig. 2A). The absence of detectable NTPD1 in the supernatant indicated that the presence of NTPD2 in the culture supernatant was not due to parasite lysis during culture, but represented active secretion (Fig. 2A). Furthermore, live cell fluorescence microscopy of promastigotes expressing NTPD2-GFP did not detect significant cell surface or intracellular fluorescence, consistent with NTPD2 being primarily a secreted protein. Interestingly, Western blot analysis detected a small pool of NTPD2-GFP within the cell pellet fraction (Fig. 2A), which is likely to represent newly synthesized NTPDase in transit to the cell surface, but below the level of detection of fluorescence microscopy. Because of the low abundance of this intracellular pool we can also not discount the possibility that NTPDase2 is directed to other intracellular organelles, such as the lysosome. In contrast, L. major promastigotes expressing NTPD1-GFP displayed a single, highly fluorescent punctate stain, at the anterior end of the parasite, proximal to the kinetoplast/flagellar pocket (Fig. 2B). This location is highly characteristic of the Golgi apparatus. L. major parasites expressing NTPD1-GFP were therefore co-transfected with a second plasmid encoding the known Golgi protein LPG1 [40] fused to mCherry. Parasites expressing both NTPD1-GFP and the Golgi marker displayed overlapping fluorescence indicative of co-localization (Fig. 2B). This co-localization was not seen in parasites transfected with either mCherry or GFP (both of which display cytoplasmic localization), indicating that NTPD1 is primarily located in the Golgi apparatus. Although yeast NTPDases have been localized to the Golgi apparatus [48], [49], this is the first time a parasite NTPDase has been identified in the Golgi apparatus, rather than being secreted from the parasite or located on the cell surface.
Figure 2. Subcellular localization of LmNTPDase-GFP fusion proteins.
A. Western blot using anti-GFP antibody demonstrating production of GFP-fusion proteins of the correct sizes by L. major parasites transfected with either pXG-NTPD1-GFP or pXG-NTPD2-GFP, and secretion of NTPD2-GFP into the culture supernatant. Lane 1: L. major + pXG-NTPD1-GFP (whole cell lysate, C), Lane 2: L. major + pXG-NTPD1-GFP culture supernatant (SN), Lane 3: L. major + pXG-NTPD2-GFP C, Lane 4, L. major + pXG-NTPD2-GFP SN. Samples were developed simultaneously on one membrane, with the vertical line representing removal of unrelated intervening lanes. B. Localization of NTPD1-GFP to the Golgi apparatus. Top panel: L. major co-transfected with pXG-NTPD1-GFP and pXG-LPG1-mCherry; middle panel: L. major co-transfected with pXG-NTPD1-GFP and pXG-SAT-mCherry; bottom panel: L. major co-transfected with pXG-/GFP+ and pXG-LPG1-mCherry. Arrow indicates co-localisation of NTPD1-GFP and LPG1-mCherry in the Golgi apparatus. Hoechst staining highlights the parasite nucleus (diffuse staining) and kinetoplast (dense staining), with the Golgi apparatus (top and bottom panel, mCherry) in the region adjacent to the kinetoplast (as expected).
NTPD1, but not NTPD2, is required for normal lesion development in mice
Previous transcript profiling studies have suggested that ntpd1 and ntpd2 are constitutively transcribed in both major developmental stages [50], [51], providing little information on potential stage-specific differences in function. To investigate the function of these enzymes we generated null mutants for each NTPDase gene, by sequential replacement of the two chromosomal alleles with drug resistance cassettes. ntpd1 was replaced with hygromycin and puromycin resistance cassettes, with gene deletion and correct integration of the resistance cassettes confirmed by triplicate PCR (S1 Fig.), demonstrating that ntpd1 is not essential under rich culture conditions. In a similar manner ntpd2 was replaced with puromycin and bleomycin cassettes, with PCR confirmation performed in triplicate (S1 Fig.), indicating that ntpd2 is also not essential in vitro. Both strains grew normally in routine culture medium.
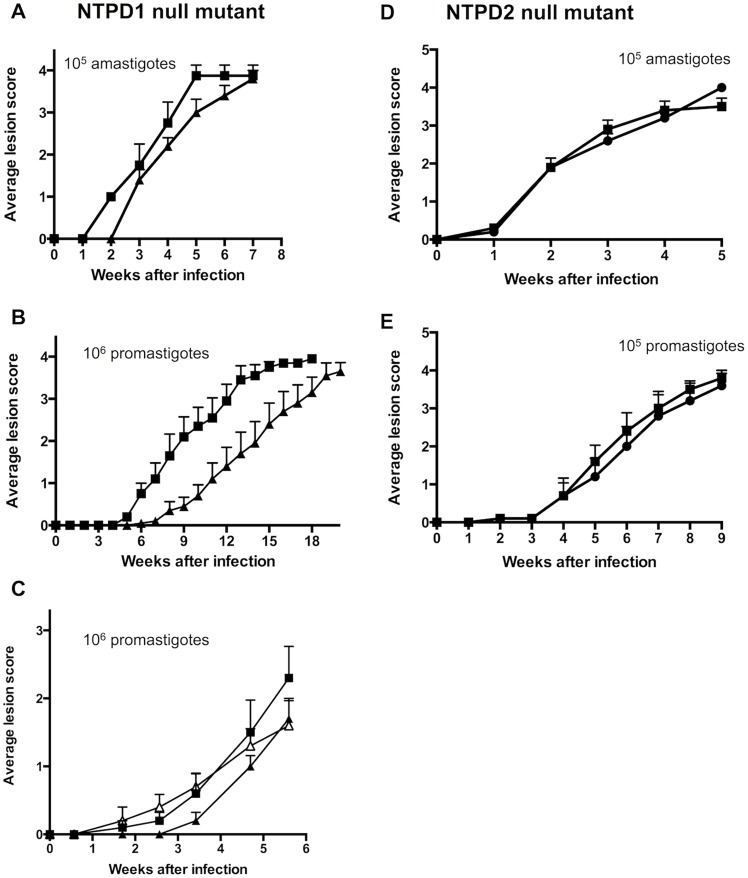
To investigate whether LmNTPDase1 or 2 is required for virulence in the mammalian host, we tested the ability of L. major Δntpd1 and Δntpd2 to induce lesions in susceptible BALB/c mice. Promastigote stages of the L. major NTPD1 null mutant exhibited a marked and highly reproducible delay in lesion development. This delay was largely abrogated by complementation of the null mutant by insertion of a full-length ntpd1 gene in the highly-transcribed ribosomal ssu locus [52]. Interestingly, no delay in lesion development was observed when amastigote stages of the NTPD1 null mutant were used to initiate the infection (Fig. 3A–C). Together, these studies demonstrate that NTPD1 is required during the early stages of promastigote infectivity, but has limited function in production of lesions following amastigote infection.
Figure 3. Subcutaneous infection of BALB/c mice with either amastigote (A, D) or promastigote (B, C, E) L. major.
A. Mice were infected with either 105 wild type L. major (squares) or 105 L. major NTPD1 null mutant (triangles) amastigotes and lesion scores monitored weekly. Error bars represent S.E.M. (n = 5). No significant difference in lesion size was observed at any time point (P>0.05, unpaired t-test). B. Mice were infected with either 106 wild type L. major (squares) or 106 L. major NTPD1 null mutant (triangles) parasites and lesion scores monitored weekly. Error bars represent S.E.M. (n = 10). Significant differences in lesion size were observed at all time points from week 6 inclusive (P<0.05, unpaired t-test). C. Mice were infected with either 106 wild type L. major + pIR1SAT (squares), 106 L. major NTPD1 null mutant + pIR1SAT (closed triangles) or 106 L. major NTPD1 null mutant + pIR1SAT-ntpd1 (open triangles). Error bars represent S.E.M. (n = 5). D and E. Mice were infected with either 105 wild type L. major (squares) or 105 L. major NTPD2 null mutant (circles) parasites and lesion scores monitored weekly. Error bars represent S.E.M. (n = 5). No significant difference in lesion size was observed between strains at any individual time point (P>0.05, two-way ANOVA).
In contrast to the NTPD1 null mutant, the NTPD2 null mutant exhibited a virulence phenotype in BALB/c mice that was indistinguishable from wild type parasites, regardless of whether promastigotes or amastigotes were used to initiate infection (Fig. 3D and 3E). Infections were repeated a number of times and it is possible that these parasites have adapted to loss of NTPD2. Regardless, these results suggest that NTPD2 is not required for virulence in the mammalian host. Lesion development within the mouse reflects both parasite replication and the host response, and our results do not rule out an alteration in parasite replication levels between wild type and the NTPD2 null mutant. However the ability to cause disease, as measured by lesion size, was unchanged between the two strains.
The L. major NTPD1 null mutant is defective in LPG elongation
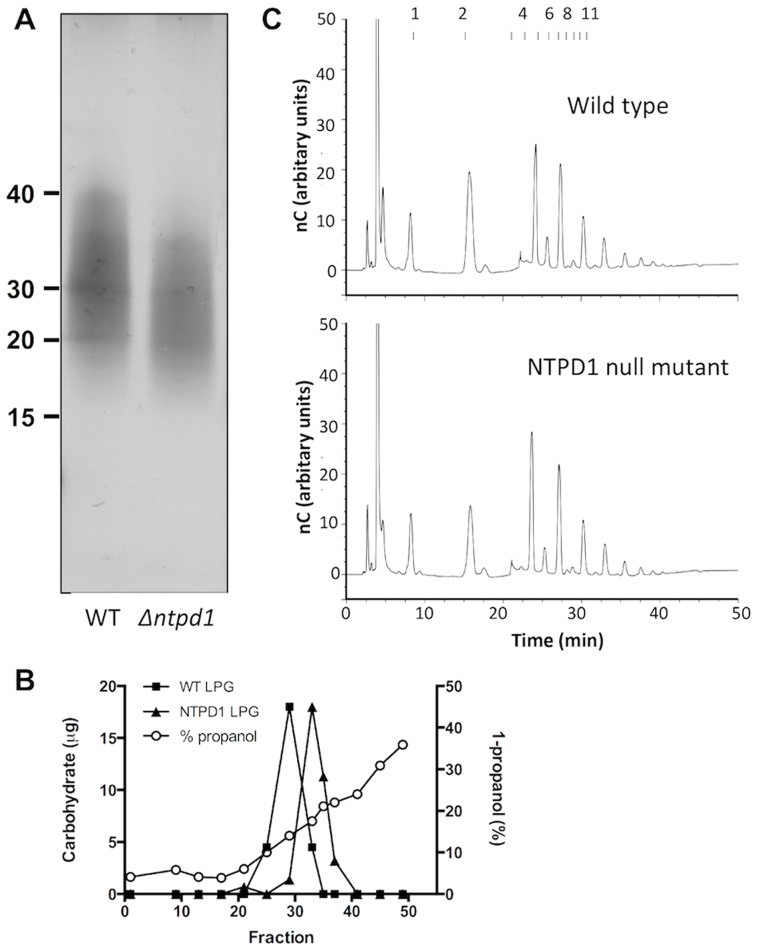
By analogy with the function of the Golgi-located yeast NTPDase, we predicted that NTPD1 may be involved in regulating the recycling of sugar-nucleotides in the Golgi lumen and hence glycosylation pathways [48], [49]. This hypothesis was further supported by the delayed lesion virulence phenotype of the NTPD1 null mutant, which is reminiscent of that seen previously for L. major mutant parasites that lack the major surface glycoconjugate, LPG [5], [53]. While LPG has multiple roles in the sandfly vector, it is only required for the early stages of promastigote infectivity in the mammalian host. LPG is not required for survival or growth of intracellular amastigotes, and LPG mutant parasites that survive the innate immune responses of the mammalian host can subsequently induce normal lesions [4], [5], as observed for the NTPD1 null mutant. To assess whether the L. major NTPD1 null mutant was defective in LPG biosynthesis, the de-lipidated wild type and mutant promastigotes were extracted in 9% 1-butanol and the lipoglycoconjugates purified by octyl-Sepharose chromatography [41]. The NTPD1 null mutant produced comparable levels of LPG as wild type parasites (Fig. 4A). As expected, both LPG preparations were visualized as smears on SDS-PAGE gels, reflecting heterogeneity in the length of the phosphoglycan chains that comprise the major portion of the LPG [42]. However, the LPG isolated from null mutant promastigotes reproducibly exhibited a lower average molecular weight on the SDS-PAGE gels (Fig. 4A) and eluted later from the octyl-Sepharose column (Fig. 4B), indicating shorter average chain length and/or reduced side chain branching. To distinguish between these possibilities, the LPG prepared from wild type and Δntpd1 promastigotes was depolymerized with mild acid treatment (40 mM TFA, 100°C, 8 min) and dephosphorylated prior to analysis by HPAEC. Both LPG preparations had essentially identical oligosaccharide repeat unit profiles (Fig. 4C). Furthermore, LC/MS analysis of the released PI lipid moieties showed that both wild type and mutant LPG contained identical very long chain (C24:0, C26:0) alkylglycerol moieties. Collectively, these structural analyses suggest that the faster SDS-PAGE mobility of LPG isolated from the NTPD1 null mutant reflects decreased phosphoglycan chain elongation, rather than altered side chain additions or increased hydrophobicity in the lipid anchor.
Figure 4. Analysis of purified LPG.
A. LPG extracted from L. major wild type (WT) and L. major Δntpd1 after SDS-PAGE and silver staining, demonstrating a clear difference in apparent molecular weight. Numbers indicate approximate molecular weight markers (kDa). B. Elution profile during octyl-Sepharose chromatography of LPG extracted from wild type L. major (squares) and the NTPD1 null mutant (triangles). LPG content was determined by orcinol staining [3:5-dihydroxy-toluene, BDH; 0.2%(w/v) in 10% H2SO4 and 50% ethanol], followed by colour development at 100°C and comparison to a known standard. The 1-propanol gradient concentration (open circles) was measured refractometrically. C. Fractionation of the dephosphorylated repeat units of LPG from wild-type and NTPD1 null mutant promastigotes. LPG was purified by octyl-Sepharose chromatography, depolymerised with 40 mM trifluoroacetic acid (8 min, 100°C) and dephosphorylated with calf intestinal alkaline phosphatase. The repeat units were desalted by passage over a mixed bed ion exchange column and chromatographed by HPAEC. The numbers at the top of the profile represent the elution positions of dextran oligomers (number of glucose units).
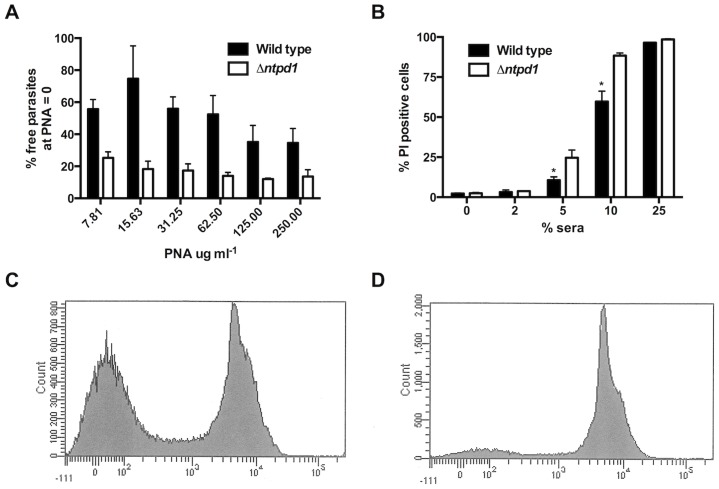
Expression of shorter LPG chains on the surface of the NTPD1 null mutant would be expected to lead to increased surface binding by the lectin, peanut agglutinin (PNA). PNA binds terminal β-Gal residues in the LPG side chains and intensity of binding is regulated by the abundance of β-Gal side chain, the extent to which these side chains are capped with arabinose and the overall length of the LPG [43]. Paradoxically, promastigotes expressing long LPG chains form surface aggregates in which LPG epitopes become cryptic and therefore bind less PNA. NTPD1 null mutant promastigotes were more effectively agglutinated than wild type promastigotes when harvested at the same stationary growth phase (Fig. 5A). Given that both wild type and mutant produce LPG with essentially identical side chain compositions (Fig. 4C), these results are consistent with the NTPD1 null promastigotes having a defect in LPG elongation.
Figure 5. Truncated LPG synthesis by L. major NTPD1 null mutants alters parasite biology.
A. The number of free-swimming parasites observed following incubation with varying concentrations of PNA, expressed as a percentage of the number of free-swimming parasites observed in the absence of PNA. Compared to wild type L. major (black columns), significantly less unbound L. major Δntpd1 (white columns) were observed at lower concentrations of PNA (*P<0.05), a trend that continued even at high concentrations of PNA. Data represents a minimum of three biological repeats. B. Percentage of parasites that were PI positive (indicating lysis) following incubation with varying concentrations of human sera. Significantly more L. major Δntpd1 (white columns) were lysed when compared to wild type L. major at sera concentrations of 5 and 10. C and D. Representative flow cytometric analysis of parasites incubated with 10% human sera, demonstrating two populations of cells (lysed and intact) for wild type L. major (C), but only one major fluorescent (lysed) cell population for L. major Δntpd1 (D). Data represents three biological repeats.
The L. major NTPD1 null mutant is more susceptible to complement lysis
To assess whether the defect in LPG chain elongation was physiologically significant, stationary phase wild type and NTPD1 null promastigotes were incubated with increasing concentrations of human serum. The complement resistance of L. major promastigotes has previously been shown to be highly dependent on LPG chain length and the formation of a thick protective surface glycocalyx [5]. NTPD1 null mutant promastigotes were significantly more sensitive to serum lysis than wild type parasites (Fig. 5B–D). In particular, FACS analysis of PI-stained parasites, showed ∼2-fold increased sensitivity at 5% serum concentrations (Fig. 5B). Collectively, these results provide strong evidence that loss of Golgi NTPDase results in less efficient elongation of LPG in virulent stationary phase promastigotes, leading to increased susceptibility to complement lysis and a marked delay in lesion development.
Discussion
The genomes of many parasitic protozoa encode one or more NTPDases, which have been implicated in various host-parasite processes [6]–[9], [19]. However, the function of these enzymes in pathogenesis has not been rigorously defined using genetic approaches. In this study we have defined the subcellular localization and function of two clearly defined NTPDase enzymes in L. major. Both proteins are predicted to contain the five ACR domains that characterize NTPDases and to be constitutively transcribed in the two major life cycle stages. Based on analysis of GFP fusion proteins, we provide evidence that NTPD1 is primarily targeted to the Golgi apparatus, while NTPD2 is secreted into the extracellular milieu. We propose that NTPD1 has an important role in regulating glycosylation pathways in the Golgi apparatus as loss of NTPD1 resulted in a defect in LPG elongation in stationary phase promastigotes. Although the overall decrease in LPG chain length in the NTPD1 null mutant was modest, it was associated with significantly increased sensitivity to complement lysis and a conspicuous delay in lesion development when promastigotes were used to initiate infection. A similar lag in lesion development was not observed when NTPD1 null mutant amastigotes were used to initiate infection, consistent with the defect being associated with a promastigote-specific virulence factor such as LPG. The similarity between the virulence phenotype of the NTPD1 null mutant and previously generated L. major LPG mutants in which assembly of the entire phosphoglycan chain has been disrupted is striking [4], [53], and strongly suggests that LPG chain elongation during stationary phase is both critical for promastigote virulence, and likely to underlie the major function of this glycoconjugate during the early stages of infection in the mammalian host.
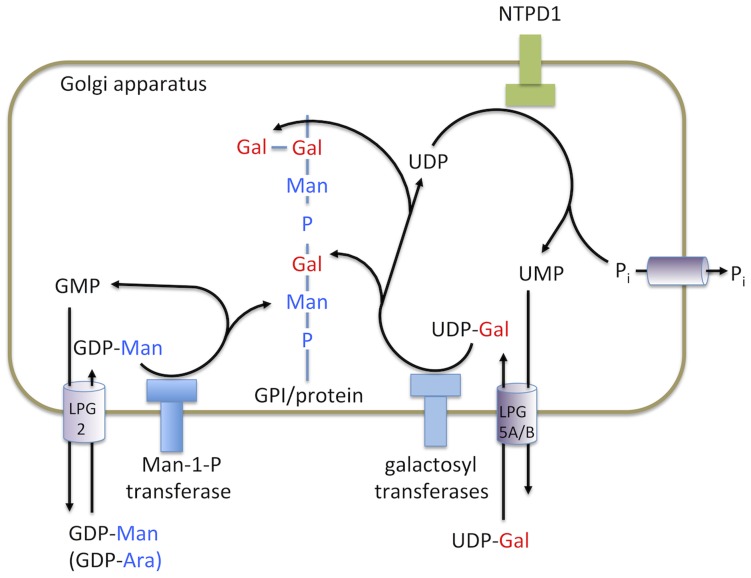
S. cerevisiae expresses two NTPDases, GDA1 and YND1, that are targeted to the Golgi apparatus with their catalytic domains orientated into the lumen [48], [49], [54]. These enzymes have been shown to hydrolyze NDP nucleotides to the corresponding NMP nucleotide, which is then used as the counter ion to import sugar nucleotides from the cytoplasm into the Golgi lumen. NTPDase-mediated hydrolysis of NDPs is thus critical for maintaining luminal levels of a range of sugar nucleotides that are used by Golgi glycosyltransferases [55]. In Leishmania, the Golgi apparatus contains enzymes required for the assembly and elongation of complex phosphoglycans on GPI anchor precursors, as well as a number of cell surface and secreted proteophosphoglycans (PPGs). All of these phosphoglycans contain the biosynthetic repeat unit, Galβ1-4Manα1-PO4, which is assembled by sequential transfer of Manα-1phosphate and galactose to the growing phosphoglycan chain by GDP-Man and UDP-Gal-dependent Golgi glycosyltransferases, respectively. The reactions catalyzed by the UDP-Gal dependent galactosyltransferases generate UDP, which would need to be converted to UMP by a NTPDase activity in order to sustain continued import of UDP-Gal into the Golgi lumen (Fig. 6). In contrast, the GDP-Man dependent Man-1-PO4-transferase(s) generate GMP, rather than GDP, and this NMP could be used to drive import of GDP-Man independent of the NTPDase activity. Thus the Golgi NTPDase is likely to be exclusively required for the galactosyltransferase-mediated reactions and not the GDP-Man-dependent Man-1-PO4 reactions. The fact that we see a specific defect in LPG chain elongation, but not in side chain modifications in the NTPDase mutant implies that β1-4-galactosyltransferase involved in assembly of the repeat unit backbone is more sensitive to depletion of UDP-Gal in the Golgi lumen than the β1-3galactosyltransferases that add additional galactose residues to the repeat unit backbone. At present, essentially nothing is known about the mechanisms that regulate LPG elongation, notwithstanding the importance of this process during the differentiation of rapidly dividing promastigotes to non-dividing, hypervirulent metacyclic promastigotes in culture and in the sandfly vector. Our findings raise the possibility that the changes in the availability of sugar nucleotides, either through changes in the activity/expression levels of Golgi membrane transporters or the luminal orientated NTPD1, could play an important role in this respect.
Figure 6. Proposed model for the role of NTPD1 in Golgi nucleotide-sugar transport and LPG synthesis.
UDP-galactose and GDP-mannose/GDP-arabinose are transported into the Golgi via transporters LPG5A/LPG5B [37] and LPG2 [65] respectively. Galactose and mannose-phosphate are cleaved for use in phosphoglycan synthesis. Following cleavage, GMP is exchanged for GDP-mannose transport into the lumen. In the case of UDP, hydrolysis to UMP is catalyzed by NTPD1, allowing efficient ongoing transport of UDP-galactose into the Golgi lumen.
In contrast to NTPD1, deletion of NTPD2 had no measurable impact on the growth of L. major promastigotes in vitro or in vivo. As NTPD2 was secreted into the medium, it is unlikely that the absence of a detectable LPG or virulence phenotype in the NTPD2 mutant reflects redundancy between the two NTPDases. One possibility is that secreted NTPDase2 is primarily required for salvage of extracellular purines. Leishmania are purine auxotrophs but express a number of surface nucleotidases, acid phosphatases, nucleotide/nucleoside/purine base transporters, as well as intracellular enzymes involved in interconverting different purine intermediates [56]. This robust network of redundant purine salvage pathways could account for the absence of a conspicuous phenotype in the NTPD2 null mutant.
A recent study has suggested that L. braziliensis LbNTPDase1 is localized on the cell surface of promastigotes [21], and that opsonization with a polyclonal antibody directed to this protein was cytotoxic. Using this antibody, the authors also suggested that LbNTPDase1 may be additionally targeted to the mitochondria, cytoplasmic vesicles, kinetoplast and nucleus. It is possible that the Leishmania NTPDase1 homologues are targeted to different subcellular localizations in a species-specific manner and perform different functions. Further work to validate the specificity of the LbNTPDase1 polyclonal antibodies and/or determination of tagged proteins would be of interest.
Previous work demonstrated variation in the level of ecto-nucleotidase activity between Leishmania species [57]. Activity in L. major was lower than that observed for L. amazonensis, which was also more virulent in the mouse model used in the study, suggesting that the role of NTPDases in the disease process could differ between species of Leishmania. However, this study did not demonstrate that the observed ecto-nucleotidase activity was linked to ntpd gene expression, and the activity may relate to other enzymes. The same study also utilised Western blot analysis, using polyclonal antibody against T. cruzi NTPDase, to detect a band corresponding to the predicted size of NTPDase1 in L. amazonensis, but failed to identify a similar band in L. major. This may be due to failure of the antibody to recognize the L. major NTPDase, but could also suggest the natural level of expression of NTPDase1 in L. major is lower. However, in light of our findings that LmNTPDase1 localises to the Golgi apparatus, it is unlikely that lower expression of LmNTPDase1 would result in lower ecto-nucleotidase activity of L. major. Future studies taking defined genetic approaches to study NTPDases in other species of Leishmania would be extremely valuable in both defining their function, and in elucidating the value of this class of enzymes as a potential therapeutic target in Leishmania.
It is also important to recognize that a number of studies have implicated general surface-located hydrolysis of ATP, ADP (and sometimes other NTPs and NDPs) in the virulence of both Leishmania and a number of other parasites [18], [19], [58]–[62]. This observed activity has often been assumed to be due to the presence of NTPDases. However, our data raise the possibility that other classes of parasite enzymes are responsible for the observed activity and play a role in pathogenesis themselves. For example, a known NTPDase inhibitor, ARL67156, only inhibits 30% of observed ecto-ATPase activity of T. cruzi [6], suggesting that investigation of other classes of enzymes would also be worthwhile. It may be that a combinatorial approach is required, and that inhibition of two or more surface enzymes could be successful in treating disease.
In conclusion, this work considerably expands our knowledge of the role of Leishmania NTPDases in host-parasite interactions. We show for the first time that parasite NTPDases can be targeted to the Golgi, and play an important role in regulating the assembly of surface virulence factors. Unexpectedly, and notwithstanding previous studies suggesting that secreted NTPDases may have essential roles in purine acquisition, and/or host or parasite purinergic signalling, loss of the secreted NTPD2 had no discernible affect on promastigote or amastigote infectivity in mice. These studies highlight the importance of exploiting genetic approaches whenever possible in investigating the function of these enzymes in host-parasite interactions.
Supporting Information
PCR confirmation of deletion of ntpd genes in L. major . A. Schematic demonstrating the location of primers used in polymerase chain reaction (PCR) analysis (see S1 Table for specific sequences). Dotted line indicates region of chromosome included in plasmid used to generate mutant. Arrows represent approximate location of primers, either upstream of this region, within the resistance (R) gene or within the specific ntpd gene. B. PCR products indicating the presence or absence of the ntpd1 gene (ntpd1) and the correct integration of the puromycin (pur) and hygromycin (hyg) cassettes onto the chromosome in place of the ntpd1 gene. Template for each reaction was either wild type L. major (W), deionised sterile water (-) or the L. major NTPD1 null mutant (M). Expected band size for the ntpd1 PCR was 1230 base pairs (bp), for the pur integration PCR was 1276 bp and for the hyg integration PCR was 1468 bp. Results clearly indicate the complete absence of the ntpd1 gene from the deletion mutant and the integration of the two resistance genes in its place, and confirm the absence of any additional alleles encoding ntpd1 in the L. major ntpd1 deletion mutant. C. Polymerase chain reaction products indicating the presence or absence of the ntpd1 gene (ntpd2) and the correct integration of the puromycin (pur) and bleocin (ble) cassettes onto the chromosome in place of the ntpd2 gene. Template for each reaction was either wild type L. major (W), deionised sterile water (-) or the L. major NTPD2 null mutant (M). “x” indicates and empty lane. Expected band size for the ntpd2 PCR was 2047 base pairs, for the pur integration PCR was 1081 base pairs and for the ble integration PCR was 1147 base pairs. Results clearly indicate the complete absence of the ntpd2 gene from the deletion mutant, the integration of the two resistance genes in its place, and confirm the absence of any additional alleles encoding ntpd2 in the L. major ntpd2 deletion mutant. PCR analysis was performed at a number of time points during culture, as well as before and after mouse infection, and typical results are presented.
(TIFF)
Primer sequences used in genetic manipulation of L. major and screening of drug resistant parasite lines for NTPD null mutants.
(DOCX)
Acknowledgments
pIR1SAT and pXG-derived plasmids were generously provided by Professor Stephen Beverley (Washington University, Kentucky). Fluorescent images were acquired using the Deltavision Elite Microscope in the Biological Optical Microscopy Platform at the University of Melbourne with the assistance of Dr. Paul McMillan. FACS data was acquired with the assistance of Dr. Desmond Ang and the mCherry template vector was a kind gift of Professor Paul Gleeson (both from the Department of Biochemistry and Molecular Biology, Bio21 Institute of Molecular Science and Biotechnology, University of Melbourne).
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was funded by the Australian National Health and Medical Research Council (NHMRC; https://www.nhmrc.gov.au). FMS was supported by an NHMRC Postdoctoral Training fellowship and MJM is an NHMRC Principal Research Fellow. This work was supported by NHMRC project grant APP1059545. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Croft SL, Olliaro P (2011) Leishmaniasis chemotherapy–challenges and opportunities. Clinical microbiology and infection: the official publication of the European Society of Clinical Microbiology and Infectious Diseases 17: 1478–1483. [DOI] [PubMed] [Google Scholar]
- 2. Murray HW, Berman JD, Davies CR, Saravia NG (2005) Advances in leishmaniasis. Lancet 366: 1561–1577. [DOI] [PubMed] [Google Scholar]
- 3. Naderer T, Vince JE, McConville MJ (2004) Surface determinants of Leishmania parasites and their role in infectivity in the mammalian host. Curr Mol Med 4: 649–665. [DOI] [PubMed] [Google Scholar]
- 4. Spath GF, Epstein L, Leader B, Singer SM, Avila HA, et al. (2000) Lipophosphoglycan is a virulence factor distinct from related glycoconjugates in the protozoan parasite Leishmania major . Proc Natl Acad Sci U S A 97: 9258–9263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Spath GF, Garraway LA, Turco SJ, Beverley SM (2003) The role(s) of lipophosphoglycan (LPG) in the establishment of Leishmania major infections in mammalian hosts. Proc Natl Acad Sci U S A 100: 9536–9541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Santos RF, Possa MA, Bastos MS, Guedes PM, Almeida MR, et al. (2009) Influence of Ecto-nucleoside triphosphate diphosphohydrolase activity on Trypanosoma cruzi infectivity and virulence. PLoS neglected tropical diseases 3: e387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mariotini-Moura C, Bastos MS, de Castro FF, Trindade ML, de Souza Vasconcellos R, et al. (2013) Trypanosoma cruzi nucleoside triphosphate diphosphohydrolase 1 (TcNTPDase-1) biochemical characterization, immunolocalization and possible role in host cell adhesion. Acta tropica 130C: 140–147. [DOI] [PubMed] [Google Scholar]
- 8. Nakaar V, Samuel BU, Ngo EO, Joiner KA (1999) Targeted reduction of nucleoside triphosphate hydrolase by antisense RNA inhibits Toxoplasma gondii proliferation. J Biol Chem 274: 5083–5087. [DOI] [PubMed] [Google Scholar]
- 9. Kikuchi T, Furuta T, Kojima S (2001) Membrane localization and demonstration of isoforms of nucleoside triphosphate hydrolase from Toxoplasma gondii . Parasitology 122 Pt 1: 15–23. [DOI] [PubMed] [Google Scholar]
- 10. Maioli TU, Takane E, Arantes RM, Fietto JL, Afonso LC (2004) Immune response induced by New World Leishmania species in C57BL/6 mice. Parasitol Res 94: 207–212. [DOI] [PubMed] [Google Scholar]
- 11. Leite PM, Gomes RS, Figueiredo AB, Serafim TD, Tafuri WL, et al. (2012) Ecto-nucleotidase activities of promastigotes from Leishmania (Viannia) braziliensis relates to parasite infectivity and disease clinical outcome. PLoS Negl Trop Dis 6: e1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. de Souza MC, de Assis EA, Gomes RS, Marques da Silva Ede A, Melo MN, et al. (2010) The influence of ecto-nucleotidases on Leishmania amazonensis infection and immune response in C57B/6 mice. Acta Trop 115: 262–269. [DOI] [PubMed] [Google Scholar]
- 13. Knowles AF (2011) The GDA1_CD39 superfamily: NTPDases with diverse functions. Purinergic Signal 7: 21–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Robson SC, Sevigny J, Zimmermann H (2006) The E-NTPDase family of ectonucleotidases: Structure function relationships and pathophysiological significance. Purinergic Signalling 2: 409–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sansom FM, Newton HJ, Crikis S, Cianciotto NP, Cowan PJ, et al. (2007) A bacterial ecto-triphosphate diphosphohydrolase similar to human CD39 is essential for intracellular multiplication of Legionella pneumophila. Cell Microbiol 9: 1922–1935. [DOI] [PubMed] [Google Scholar]
- 16. Sansom FM, Riedmaier P, Newton HJ, Dunstone MA, Muller CE, et al. (2008) Enzymatic properties of an ecto-nucleoside triphosphate diphosphohydrolase from Legionella pneumophila: substrate specificity and requirement for virulence. J Biol Chem 283: 12909–12918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Meyer-Fernandes JR, Dutra PM, Rodrigues CO, Saad-Nehme J, Lopes AH (1997) Mg-dependent ecto-ATPase activity in Leishmania tropica . Arch Biochem Biophys 341: 40–46. [DOI] [PubMed] [Google Scholar]
- 18. Berredo-Pinho M, Peres-Sampaio CE, Chrispim PP, Belmont-Firpo R, Lemos AP, et al. (2001) A Mg-dependent ecto-ATPase in Leishmania amazonensis and its possible role in adenosine acquisition and virulence. Arch Biochem Biophys 391: 16–24. [DOI] [PubMed] [Google Scholar]
- 19. Pinheiro CM, Martins-Duarte ES, Ferraro RB, Fonseca de Souza AL, Gomes MT, et al. (2006) Leishmania amazonensis: Biological and biochemical characterization of ecto-nucleoside triphosphate diphosphohydrolase activities. Experimental parasitology 114: 16–25. [DOI] [PubMed] [Google Scholar]
- 20. Ennes-Vidal V, Castro RO, Britto C, Barrabin H, D'Avila-Levy CM, et al. (2011) CrATP interferes in the promastigote-macrophage interaction in Leishmania amazonensis infection. Parasitology 138: 960–968. [DOI] [PubMed] [Google Scholar]
- 21. Porcino GN, Carvalho-Campos C, Maia AC, Detoni ML, Faria-Pinto P, et al. (2012) Leishmania (Viannia) braziliensis nucleoside triphosphate diphosphohydrolase (NTPDase 1): localization and in vitro inhibition of promastigotes growth by polyclonal antibodies. Exp Parasitol 132: 293–299. [DOI] [PubMed] [Google Scholar]
- 22. Deaglio S, Dwyer KM, Gao W, Friedman D, Usheva A, et al. (2007) Adenosine generation catalyzed by CD39 and CD73 expressed on regulatory T cells mediates immune suppression. J Exp Med 204: 1257–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Borsellino G, Kleinewietfeld M, Di Mitri D, Sternjak A, Diamantini A, et al. (2007) Expression of ectonucleotidase CD39 by Foxp3+ Treg cells: hydrolysis of extracellular ATP and immune suppression. Blood 110: 1225–1232. [DOI] [PubMed] [Google Scholar]
- 24. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215: 403–410. [DOI] [PubMed] [Google Scholar]
- 25. Sansom FM (2012) The role of the NTPDase enzyme family in parasites: what do we know, and where to from here? Parasitology 139: 963–980. [DOI] [PubMed] [Google Scholar]
- 26. Sansom FM, Robson SC, Hartland EL (2008) Possible effects of microbial ecto-nucleoside triphosphate diphosphohydrolases on host-pathogen interactions. Microbiol Mol Biol Rev 72: 765–781 Table of Contents.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Goujon M, McWilliam H, Li W, Valentin F, Squizzato S, et al. (2010) A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res 38: W695–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, et al. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948. [DOI] [PubMed] [Google Scholar]
- 29. Letunic I, Copley RR, Pils B, Pinkert S, Schultz J, et al. (2006) SMART 5: domains in the context of genomes and networks. Nucleic Acids Res 34: D257–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schultz J, Milpetz F, Bork P, Ponting CP (1998) SMART, a simple modular architecture research tool: identification of signaling domains. Proc Natl Acad Sci U S A 95: 5857–5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sansom FM, Tang L, Ralton JE, Saunders EC, Naderer T, et al. (2013) Leishmania major methionine sulfoxide reductase A is required for resistance to oxidative stress and efficient replication in macrophages. PloS one 8: e56064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cruz A, Coburn CM, Beverley SM (1991) Double targeted gene replacement for creating null mutants. Proc Natl Acad Sci U S A 88: 7170–7174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Naderer T, Ellis MA, Sernee MF, De Souza DP, Curtis J, et al. (2006) Virulence of Leishmania major in macrophages and mice requires the gluconeogenic enzyme fructose-1,6-bisphosphatase. Proc Natl Acad Sci U S A 103: 5502–5507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Freedman DJ, Beverley SM (1993) Two more independent selectable markers for stable transfection of Leishmania . Molecular and biochemical parasitology 62: 37–44. [DOI] [PubMed] [Google Scholar]
- 35. Naderer T, Wee E, McConville MJ (2008) Role of hexosamine biosynthesis in Leishmania growth and virulence. Molecular microbiology 69: 858–869. [DOI] [PubMed] [Google Scholar]
- 36. Robinson KA, Beverley SM (2003) Improvements in transfection efficiency and tests of RNA interference (RNAi) approaches in the protozoan parasite Leishmania . Mol Biochem Parasitol 128: 217–228. [DOI] [PubMed] [Google Scholar]
- 37. Capul AA, Barron T, Dobson DE, Turco SJ, Beverley SM (2007) Two functionally divergent UDP-Gal nucleotide sugar transporters participate in phosphoglycan synthesis in Leishmania major . J Biol Chem 282: 14006–14017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ha DS, Schwarz JK, Turco SJ, Beverley SM (1996) Use of the green fluorescent protein as a marker in transfected Leishmania . Mol Biochem Parasitol 77: 57–64. [DOI] [PubMed] [Google Scholar]
- 39. Houghton FJ, Bellingham SA, Hill AF, Bourges D, Ang DKY, et al. (2012) Arl5b is a Golgi-localised small G protein involved in the regulation of retrograde transport. Experimental Cell Research 318: 464–477. [DOI] [PubMed] [Google Scholar]
- 40. Zhang K, Barron T, Turco SJ, Beverley SM (2004) The LPG1 gene family of Leishmania major . Mol Biochem Parasitol 136: 11–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. McConville MJ, Bacic A, Mitchell GF, Handman E (1987) Lipophosphoglycan of Leishmania major that vaccinates against cutaneous leishmaniasis contains an alkylglycerophosphoinositol lipid anchor. Proc Natl Acad Sci U S A 84: 8941–8945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. McConville MJ, Thomas-Oates JE, Ferguson MA, Homans SW (1990) Structure of the lipophosphoglycan from Leishmania major . J Biol Chem 265: 19611–19623. [PubMed] [Google Scholar]
- 43. Sacks DL, Pimenta PF, McConville MJ, Schneider P, Turco SJ (1995) Stage-specific binding of Leishmania donovani to the sand fly vector midgut is regulated by conformational changes in the abundant surface lipophosphoglycan. J Exp Med 181: 685–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Titus RG, Marchand M, Boon T, Louis JA (1985) A limiting dilution assay for quantifying Leishmania major in tissues of infected mice. Parasite immunology 7: 545–555. [DOI] [PubMed] [Google Scholar]
- 45. Ivens AC, Peacock CS, Worthey EA, Murphy L, Aggarwal G, et al. (2005) The genome of the kinetoplastid parasite, Leishmania major. Science 309: 436–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Peacock CS, Seeger K, Harris D, Murphy L, Ruiz JC, et al. (2007) Comparative genomic analysis of three Leishmania species that cause diverse human disease. Nat Genet 39: 839–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kirley TL, Crawford PA, Smith TM (2006) The structure of the nucleoside triphosphate diphosphohydrolases (NTPDases) as revealed by mutagenic and computational modeling analyses. Purinergic Signal 2: 379–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gao XD, Kaigorodov V, Jigami Y (1999) YND1, a homologue of GDA1, encodes membrane-bound apyrase required for Golgi N- and O-glycosylation in Saccharomyces cerevisiae . J Biol Chem 274: 21450–21456. [DOI] [PubMed] [Google Scholar]
- 49. Abeijon C, Yanagisawa K, Mandon EC, Hausler A, Moremen K, et al. (1993) Guanosine diphosphatase is required for protein and sphingolipid glycosylation in the Golgi lumen of Saccharomyces cerevisiae . The Journal of cell biology 122: 307–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Leifso K, Cohen-Freue G, Dogra N, Murray A, McMaster WR (2007) Genomic and proteomic expression analysis of Leishmania promastigote and amastigote life stages: the Leishmania genome is constitutively expressed. Mol Biochem Parasitol 152: 35–46. [DOI] [PubMed] [Google Scholar]
- 51. Rochette A, Raymond F, Ubeda JM, Smith M, Messier N, et al. (2008) Genome-wide gene expression profiling analysis of Leishmania major and Leishmania infantum developmental stages reveals substantial differences between the two species. BMC Genomics 9: 255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Misslitz A, Mottram JC, Overath P, Aebischer T (2000) Targeted integration into a rRNA locus results in uniform and high level expression of transgenes in Leishmania amastigotes . Mol Biochem Parasitol 107: 251–261. [DOI] [PubMed] [Google Scholar]
- 53. Capul AA, Hickerson S, Barron T, Turco SJ, Beverley SM (2007) Comparisons of mutants lacking the Golgi UDP-galactose or GDP-mannose transporters establish that phosphoglycans are important for promastigote but not amastigote virulence in Leishmania major . Infect Immun 75: 4629–4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Abeijon C, Orlean P, Robbins PW, Hirschberg CB (1989) Topography of glycosylation in yeast: characterization of GDPmannose transport and lumenal guanosine diphosphatase activities in Golgi-like vesicles. Proc Natl Acad Sci U S A 86: 6935–6939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Liu L, Xu YX, Hirschberg CB (2010) The role of nucleotide sugar transporters in development of eukaryotes. Semin Cell Dev Biol 21: 600–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Boitz JM, Ullman B (2013) Adenine and adenosine salvage in Leishmania donovani . Mol Biochem Parasitol 190: 51–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. de Almeida Marques-da-Silva E, de Oliveira JC, Figueiredo AB, de Souza Lima Junior D, Carneiro CM, et al. (2008) Extracellular nucleotide metabolism in Leishmania: influence of adenosine in the establishment of infection. Microbes and infection/Institut Pasteur 10: 850–857. [DOI] [PubMed] [Google Scholar]
- 58. de Jesus JB, de Sa Pinheiro AA, Lopes AH, Meyer-Fernandes JR (2002) An ectonucleotide ATP-diphosphohydrolase activity in Trichomonas vaginalis stimulated by galactose and its possible role in virulence. Z Naturforsch [C] 57: 890–896. [DOI] [PubMed] [Google Scholar]
- 59. Peres-Sampaio CE, de Almeida-Amaral EE, Giarola NL, Meyer-Fernandes JR (2008) Leishmania amazonensis: effects of heat shock on ecto-ATPase activity. Experimental parasitology 119: 135–143. [DOI] [PubMed] [Google Scholar]
- 60. Tasca T, Bonan CD, De Carli GA, Sarkis JJ, Alderete JF (2005) Heterogeneity in extracellular nucleotide hydrolysis among clinical isolates of Trichomonas vaginalis . Parasitology 131: 71–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Bisaggio DF, Peres-Sampaio CE, Meyer-Fernandes JR, Souto-Padron T (2003) Ecto-ATPase activity on the surface of Trypanosoma cruzi and its possible role in the parasite-host cell interaction. Parasitol Res 91: 273–282. [DOI] [PubMed] [Google Scholar]
- 62. Meyer-Fernandes JR, Saad-Nehme J, Peres-Sampaio CE, Belmont-Firpo R, Bisaggio DF, et al. (2004) A Mg-dependent ecto-ATPase is increased in the infective stages of Trypanosoma cruzi . Parasitol Res 93: 41–50. [DOI] [PubMed] [Google Scholar]
- 63. Letunic I, Bork P (2007) Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics 23: 127–128. [DOI] [PubMed] [Google Scholar]
- 64. Letunic I, Bork P (2011) Interactive Tree Of Life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res 39: W475–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Ma D, Russell DG, Beverley SM, Turco SJ (1997) Golgi GDP-mannose uptake requires Leishmania LPG2. A member of a eukaryotic family of putative nucleotide-sugar transporters. J Biol Chem 272: 3799–3805. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PCR confirmation of deletion of ntpd genes in L. major . A. Schematic demonstrating the location of primers used in polymerase chain reaction (PCR) analysis (see S1 Table for specific sequences). Dotted line indicates region of chromosome included in plasmid used to generate mutant. Arrows represent approximate location of primers, either upstream of this region, within the resistance (R) gene or within the specific ntpd gene. B. PCR products indicating the presence or absence of the ntpd1 gene (ntpd1) and the correct integration of the puromycin (pur) and hygromycin (hyg) cassettes onto the chromosome in place of the ntpd1 gene. Template for each reaction was either wild type L. major (W), deionised sterile water (-) or the L. major NTPD1 null mutant (M). Expected band size for the ntpd1 PCR was 1230 base pairs (bp), for the pur integration PCR was 1276 bp and for the hyg integration PCR was 1468 bp. Results clearly indicate the complete absence of the ntpd1 gene from the deletion mutant and the integration of the two resistance genes in its place, and confirm the absence of any additional alleles encoding ntpd1 in the L. major ntpd1 deletion mutant. C. Polymerase chain reaction products indicating the presence or absence of the ntpd1 gene (ntpd2) and the correct integration of the puromycin (pur) and bleocin (ble) cassettes onto the chromosome in place of the ntpd2 gene. Template for each reaction was either wild type L. major (W), deionised sterile water (-) or the L. major NTPD2 null mutant (M). “x” indicates and empty lane. Expected band size for the ntpd2 PCR was 2047 base pairs, for the pur integration PCR was 1081 base pairs and for the ble integration PCR was 1147 base pairs. Results clearly indicate the complete absence of the ntpd2 gene from the deletion mutant, the integration of the two resistance genes in its place, and confirm the absence of any additional alleles encoding ntpd2 in the L. major ntpd2 deletion mutant. PCR analysis was performed at a number of time points during culture, as well as before and after mouse infection, and typical results are presented.
(TIFF)
Primer sequences used in genetic manipulation of L. major and screening of drug resistant parasite lines for NTPD null mutants.
(DOCX)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.