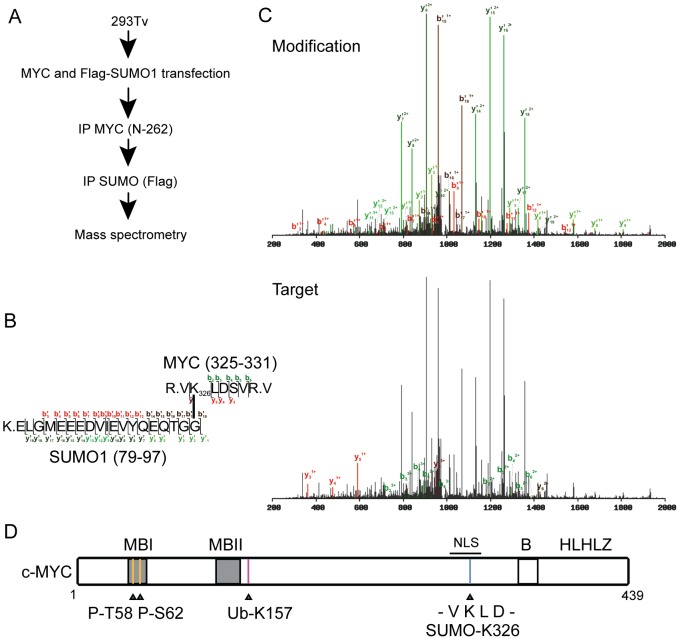
Figure 2. Identification of a SUMO site on MYC.
(A) 293Tv cells were transfected with MYC and Flag-SUMO1. Cells were then lysed under denaturing conditions and sequential pulldowns were performed first for MYC and then for FlagSUMO1. The resulting eluate was subjected to mass spectrometry analysis. (B) Observed sequence of the detected peptide fragments for both the modification (SUMO1) and the target (MYC). (C) Mass spectrometry fragmentation spectra for MYC (target) and SUMO (modification) demonstrating MYC SUMOylation at K326. (D) Diagrammatic representation of MYC and the post-translational modifications observed by mass spectrometry. P: phosphorylation, Ub: ubiquitylation, B: basic region, HLH-LZ: helix-loop-helix leucine zipper, NLS: nuclear localization signal. Grey boxes indicate regions of homology among MYC family proteins, termed MYC Boxes (MBs).