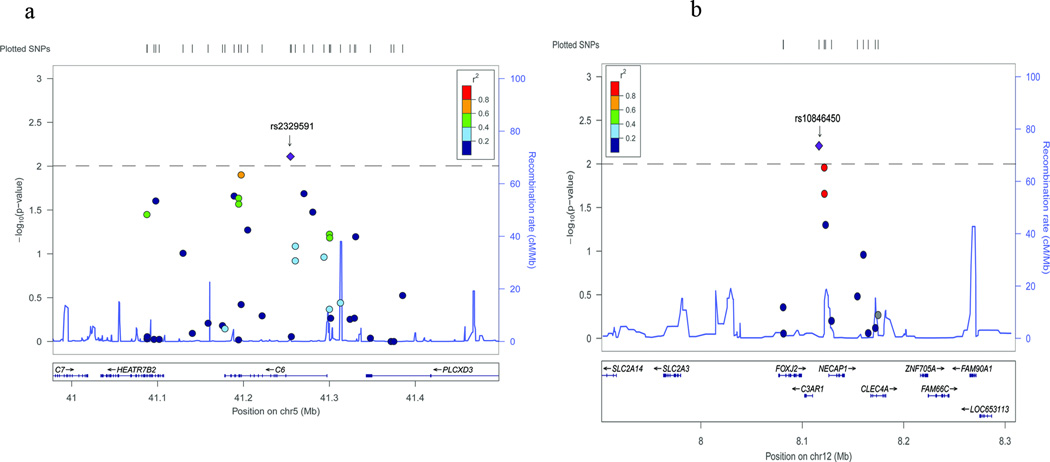
Figure 1.
Analysis of SNPs in and close to C3AR1 (a) and C6 (b) for association with CAD in Chinese Han GeneID populations using GWAS data. Regional association plots of the GWAS data are shown for C3AR1 (a) and C6 (b). Each panel shows SNPs plotted by their positions on the corresponding chromosome against −log10 P. Estimated recombination rates from 1000 genomes (JPT+CHB populations) were plotted in blue to reflect the local linkage disequilibrium (LD) structure on a secondary y axis. The most significant lead SNP (diamond) is annotated with its observed P value, and flanking SNPs (circle) are color-coded to represent the pairwise r2 measure of LD with the lead SNP: red, r2 ≥ 0·8; orange, 0·6 ≤ r2 < 0·8; green, 0·4 ≤ r2<0·6; light blue, 0·2 ≤ r2< 0·4; blue, r2 < 0·2. All positions are based on hg18. These plots were generated by Locuszoom (https://statgen.sph.umich.edu/locuszoom/)