Abstract
Cellular uptake of vitamin A, production of visual chromophore, and triglyceride homeostasis in adipocytes depend on two representatives of the vertebrate N1pC/P60 protein family, lecithin:retinol acyltransferase (LRAT) and HRAS-like tumor suppressor 3 (HRASLS3). Both proteins function as lipid-metabolizing enzymes but differ in their substrate preferences and dominant catalytic activity. The mechanism of this catalytic diversity is not understood. Here, by using a gain-of-function approach, we identified a specific sequence responsible for the substrate specificity of N1pC/P60 proteins. A 2.2 Å crystal structure of HRASLS3/LRAT chimeric enzyme in a thioester catalytic intermediate state revealed a major structural rearrangement accompanied by 3D-domain swapping dimerization not observed in native HRASLS proteins. Structural changes affecting the active site environment contributed to slower hydrolysis of the catalytic intermediate supporting efficient acyl transfer. These findings reveal structural adaption that facilitates selective catalysis and mechanism responsible for diverse substrate specificity within the LRAT-like enzyme family.
Keywords: acyltransferase, LRAT, vitamin A, retinol, HRAS-like tumor suppressor
INTRODUCTION
Enzymatic catalysis at the phospholipid membrane interface is vital for numerous fundamental biological processes including lipid metabolism and signaling1,2. Many membrane enzymes are interfacial and access substrate directly from a lipid-water interface. Well-recognized enzymatic activities requiring membrane protein interactions include phospholipid hydrolysis and phospholipid-dependent acyl transfer. Both are often mediated by a catalytic mechanism derived from serine or cysteine proteases3,4. Yet, the molecular basis for functional diversification and adaptation to alternative substrates by similar but distinct enzymatic mechanisms is incompletely understood. The main challenge in mechanistic and structural studies of membrane enzymes is the complicated nature of the lipid-enzyme interactions and their relationship to substrate binding and processing. Physiologically important members of the vertebrate N1pC/P60 protein family constitute excellent models to investigate the fundamental molecular mechanisms that govern interfacial catalysis.
In vertebrates, members of the N1pC/P60 multigene family of integral membrane proteins perform catalysis at the lipid membrane-water interface with phospholipids employed as substrates5. The human genome encodes six proteins that belong to this class of enzymes including lecithin:retinol acyltransferase (LRAT) and closely related HRAS-like tumor suppressors (HRASLS1–5), also referred to as LRAT-like proteins5,6. LRAT is the main enzyme that catalyzes vitamin A (all-trans-retinol) esterification7,8. Consequently, lack of LRAT activity causes poor retention of all-trans-retinol in peripheral tissues including the retinal pigmented epithelium (RPE), which leads to vitamin A deficiency and blindness7,9,10. HRASLS3 (also known as adipose-specific phospholipase or AdPLA) plays a dominant role in regulating lipolysis in white adipose tissue11. By controlling production of prostaglandin E2, HRASLS3 indirectly affects lipid metabolism by decreasing lipolysis and promoting triacylglycerol accumulation. Deletion of HRASLS3 prevents weight gain in both mice fed with a high caloric diet and genetically obese animals lacking leptin11. The dominant physiological role of HRASLS3 in lipid metabolism makes this enzyme an attractive target for pharmacologic treatment of obesity and the related metabolic syndrome12.
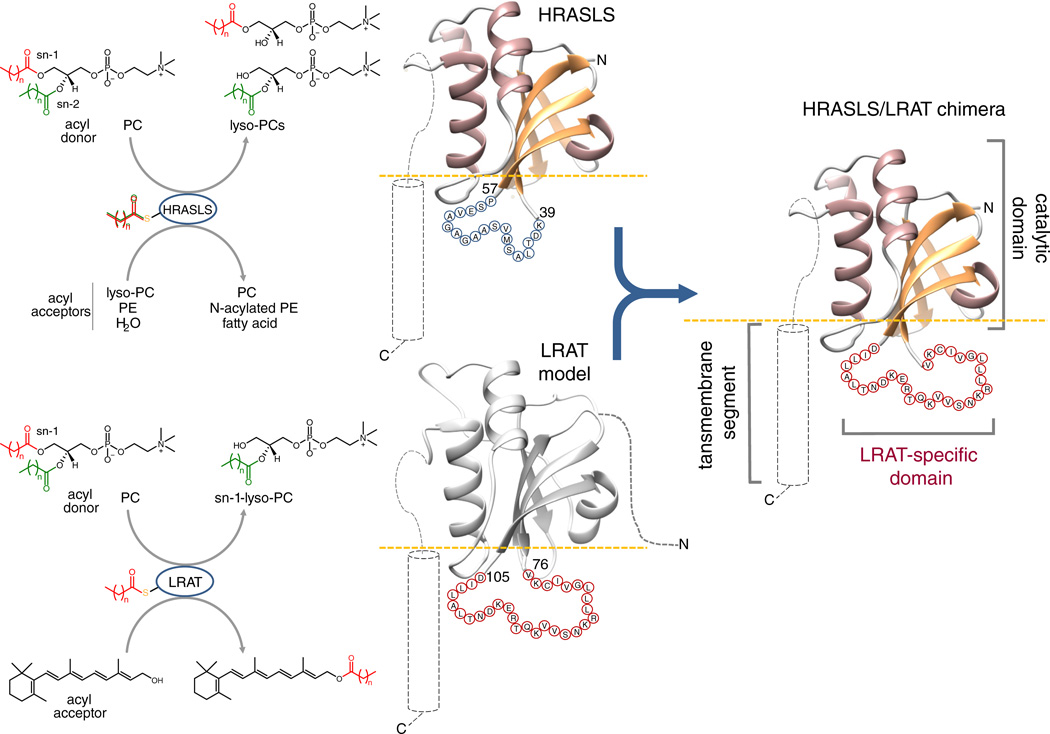
LRAT-like proteins share a common catalytic domain fold and an unconventional active site featuring a Cys-His-His catalytic triad13. Despite having originated from cell-wall N1pC/P60 peptidases,5 LRAT-like enzymes evolved alternative to peptidases activity11,13–16. LRAT, a dominant acyltransferase enzyme, catalyzes the formation of retinyl esters by transferring an acyl group from the sn-1 position of phosphatidylcholine (PC) to vitamin A17. In contrast, HRASLS enzymes function as phospholipases A1/2, cleaving PC and phosphatidylethanolamine (PE) to generate the corresponding lyso-phospholipids and free fatty acids14,18. These two enzymes also catalyze both N-acylation of PE to form N-acyl-PE and O-acylation of lyso-PC to form PC13–15,18. Despite high sequence conservation in catalytic domain regions of HRASLS proteins and LRAT, the former cannot utilize vitamin A as an acyl acceptor and lack specificity towards the sn-1 ester cleavage site13 (Fig. 1). These fundamental alterations in the enzymatic activities of closely related proteins do not arise from differences in their catalytic mechanisms,4,13,16,19 raising the question of how nature generates enzymatic diversity from similar structural elements.
Figure 1. Design of HRASLS/LRAT chimeric proteins.
Although both HRASLS enzymes and LRAT utilize phospholipids as acyl donors, they differ as to the regio-selectivity of ester bond cleavage and acyl acceptor specificity. To construct HRASLS/LRAT chimeric enzymes, the 19-aa loop connecting β-strands 3 and 4 in native HRASLS proteins (aa 41 – 57) was replaced with the 30-aa LRAT-specific sequence (aa 78 – 106 in mouse LRAT). The involved protein segments are shown as sequences of blue and red circles, each with the single letter aa code.
To delineate the mechanistic aspects of interfacial catalysis in vertebrate N1pC/P60 enzymes, we identified a 30-amino acid (aa) sequence responsible for acquisition of LRAT enzymatic activity by the HRASLS proteins. Next, we report the 2.2 Å crystal structure of the HRASLS3/LRAT chimeric protein in an acyl-thioester catalytic intermediate state that provides a mechanistic explanation for the altered enzymatic activity. We found that this 30-aa insertion differentiates LRAT from HRASLS proteins by inducing a structural rearrangement associated with domain-swapping dimerization. Evidence is provided that these structural changes directly affect rates of hydrolysis of the thioester catalytic intermediate, enabling effective acyl transferase activity. Thus LRAT-like proteins provide a general mechanism underlying the specificity of enzymes that operate at the membrane/water interface. Moreover, the crystallographic data reported herein represents the first high resolution structural information that sheds light on molecular architecture of LRAT.
RESULTS
LRAT-specific sequence changes activity of HRASLS enzymes
Determination of crystal structures of HRASLS213 and HRASLS313,20 allowed a re-interpretation of the sequence alignment of vertebrate LRAT-like protein family members in the context of their structure and membrane topology (Supplementary Results, Supplementary Fig. 1). LRAT-like proteins revealed an overall common molecular architecture featuring a conserved catalytic domain and a hydrophobic membrane-anchoring region predicted to be a single transmembrane α-helix at the C-terminus. A distinctive feature of the LRAT sequence is an 11-aa insertion within the catalytic domain followed by a 19-aa stretch conserved among LRATs but not homologous with the sequences of other members of this enzyme class (Supplementary Fig. 1 and 2). Based on the HRASLS proteins’ crystal structures, this 30-aa long domain forms an extended linker between the β3 and β4 strands of the catalytic core of the enzyme (Fig. 1). The hydrophobic nature and overall position of this segment with respect to the lipid membrane suggests that it may be directly involved in the protein-lipid interaction and thus determine the interphase catalytic properties of the enzyme. To test this hypothesis, we took advantage of the high homology between HRASLS proteins and LRAT as well as their common catalytic strategy4,13 to design, heterologously express, and purify three chimeric proteins (HRASLS2/LRAT, HRASLS3/LRAT, and HRASLS4/LRAT) in which the native linkers between β-strands 3 and 4 of the catalytic domains of HRASLS2, 3, and 4 were replaced by the 30-aa sequence characteristic of mouse LRAT (Fig. 1; Supplementary Fig. 3).
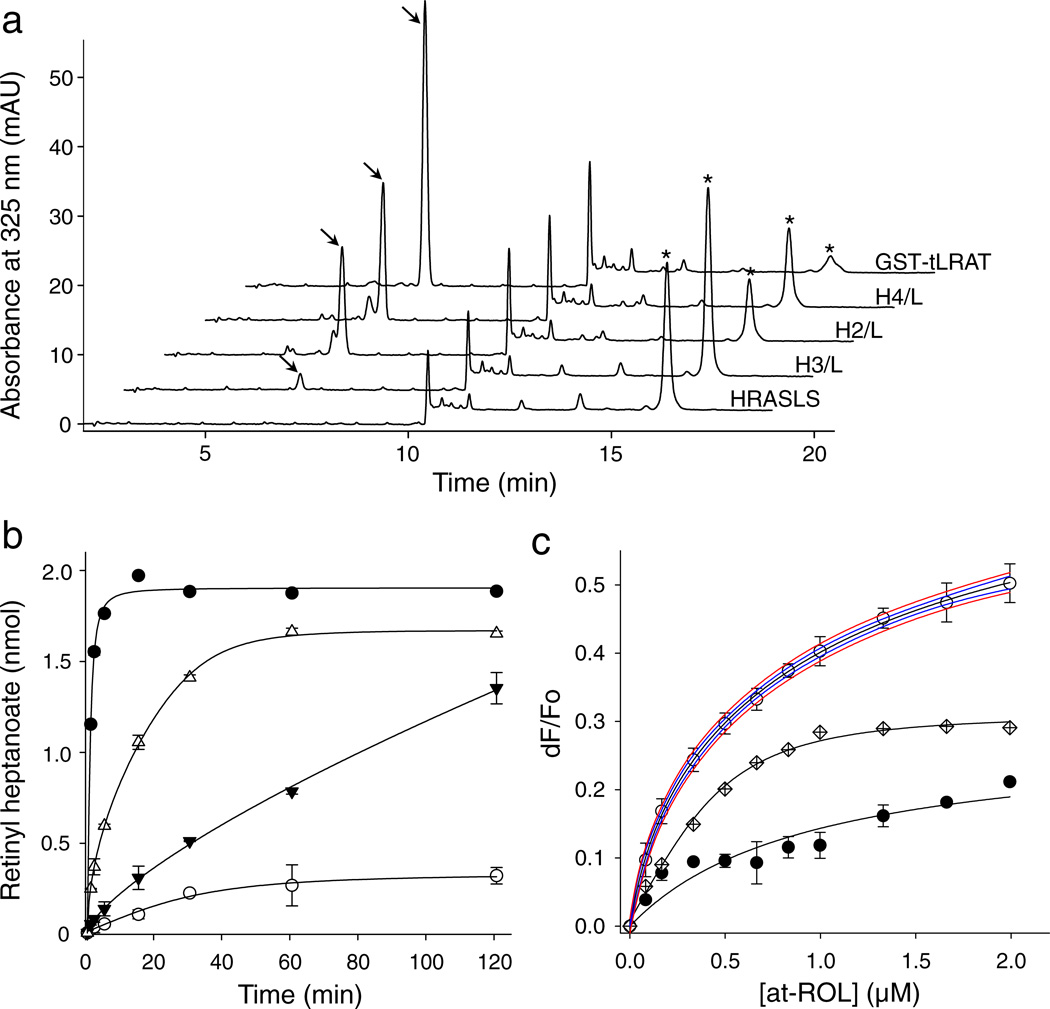
In contrast to LRAT, none of the native HRASLS enzymes accepted vitamin A as a substrate to catalyze retinyl ester formation (Fig. 2a). But strikingly, all of the examined purified HRASLS/LRAT chimeric proteins revealed robust retinyl ester production in a standard enzymatic assay (Fig.2a and 2b). Although the efficiency of the reaction diverged significantly between the chimeras as evidenced by their Vmax values, their apparent Michaelis constants (KM) were identical to those determined for LRAT (Supplementary Fig. 4). Importantly, the efficiency of retinyl ester production by the chimeric enzymes was proportional to the rates of PC hydrolysis determined for native HRASLS proteins under comparable experimental conditions13. Processing of vitamin A requires interaction of an enzyme with this retinoid substrate. Binding of all-trans-retinol was examined by fluorescence methodology and the affinity of this interaction was calculated based on the changes in internal fluorescence of a single Trp residue upon titration with the ligand. Unlike native HRASLS enzymes, the chimeric proteins gained the ability to bind vitamin A with sub-micromolar affinities similar to that calculated for LRAT (Fig. 2c; Supplementary Fig. 4). In a reverse experiment, the 30-aa linker between β3 and β4 strands of LRAT was trimmed to leave only a short sequence of polar residues (NDKERTQK) sufficient to ensure structural integrity. This altered protein retained the ability to hydrolyze PC but did not generate retinyl esters from all-trans-retinol (Supplementary Fig. 5). Together these results demonstrate the critical role of the 30-aa linker sequence for the LRAT activity of this enzyme.
Figure 2. HRASLS/LRAT chimeras catalyze formation of retinyl esters.
(a) HPLC separation of retinoids extracted after a 1 h incubation of all-trans-retinol (20 µM) and 7:0,7:0-PC (1 mM) with the tested enzymes. Peaks that correspond to retinyl heptanoate and all-trans-retinol are marked with arrows and asterisks, respectively. The chromatogram labeled as HRASLS is representative for HRASLS2, 3, and 4. (b) Comparison of the time courses of retinyl ester formation for LRAT and HRASLS/LRAT. Symbols correspond to GST-tLRAT (●), HRASLS4/LRAT (Δ), HRASLS2/LRAT (▼), and HRASLS3/LAT (○). (c) Binding of all-trans-retinol by HRASLS3/LRAT. Interaction of the retinoid ligand with HRASLS3/LRAT (○) was determined by monitoring the quenching of internal fluorescence from a single Trp residue present in the protein sequence. Blue and red lines represent the 95% confidence intervals and prediction bands, respectively, for data fitted with a two site saturation ligand binding model for specific and nonspecific ligand-protein interactions. Data obtained for native HRASLS3 (●) was used as a control for non-specific binding. Points shown with crossed diamonds correspond to the specific binding component obtained by subtracting the HRASLS3 signal from the HRASLS3/LRAT data. Kd values for all-trans-retinol binding obtained from all tested chimeric proteins are listed in Supplemental Fig. 4. All experiments were repeated three times in triplicate. Data are presented as mean values ± s.d.
In addition to vitamin A processing, a characteristic property that differentiates LRAT from HRASLS proteins is its absolute specificity towards sn-1 phospholipid ester cleavage17,21. Thus, we next examined the positional specificity of phospholipid cleavage with native and chimeric proteins by using the sn-2 chromophore-labeled lipid substrate, 1-hexanoyl-2-(6-((7-nitro-2-1,3-benzoxadiazol-4-yl)amino)hexanoyl)-sn-glycero-3-phosphocholine (NBD-PC)13,22,23. Identification of the enzymatic reaction products by liquid chromatography-mass spectrometry (LC/MS) and their subsequent quantification based on absorbance at 450 nm revealed that HRASLS2, 3, and 4 did not exhibit regio-specificity with respect to phospholipid cleavage, consistent with previous results13,14,24,25, (Supplementary Fig. 6 and 7). However, samples incubated with the corresponding chimeric enzymes contained predominantly products of sn-1 cleavage indicating a dramatic shift in these enzymes’ specificity. Importantly, deletion of the linker between β3 and β4 strands in LRAT led to a loss of regio-specificity of acyl cleavage (Supplementary Fig. 6 and 7). Thus, incorporation of the LRAT-specific domain into the sequences of HRASLS enzymes altered the phospholipid substrate interaction as well as the overall ester bond cleavage specificity from mixed sn-1, sn-2 site to a nearly exclusive sn-1 site.
Together, these results demonstrate that it is feasible to gain lecitihin:retinol acyltransferase activity by HRASLS proteins by replacing a well-defined portion of these enzymes with a LRAT-specific sequence. Therefore, by using a gain-of-function approach, we identified a 30-aa LRAT domain responsible for this enzyme’s regio-specificity for phospholipid cleavage and ability to catalyze all-trans-retinol esterification.
Structural rearrangement induced by LRAT-specific domain
To gain a mechanistic insight into the molecular adaptations that led to the dramatic change in the enzymatic specificity of these chimeric proteins, we determined a 2.2 Å crystal structure for the HRASLS3/LRAT chimeric enzyme in the presence of 7:0,7:0-PC, a phospholipid substrate (Supplementary Table 1 and Supplemental Fig. 8).
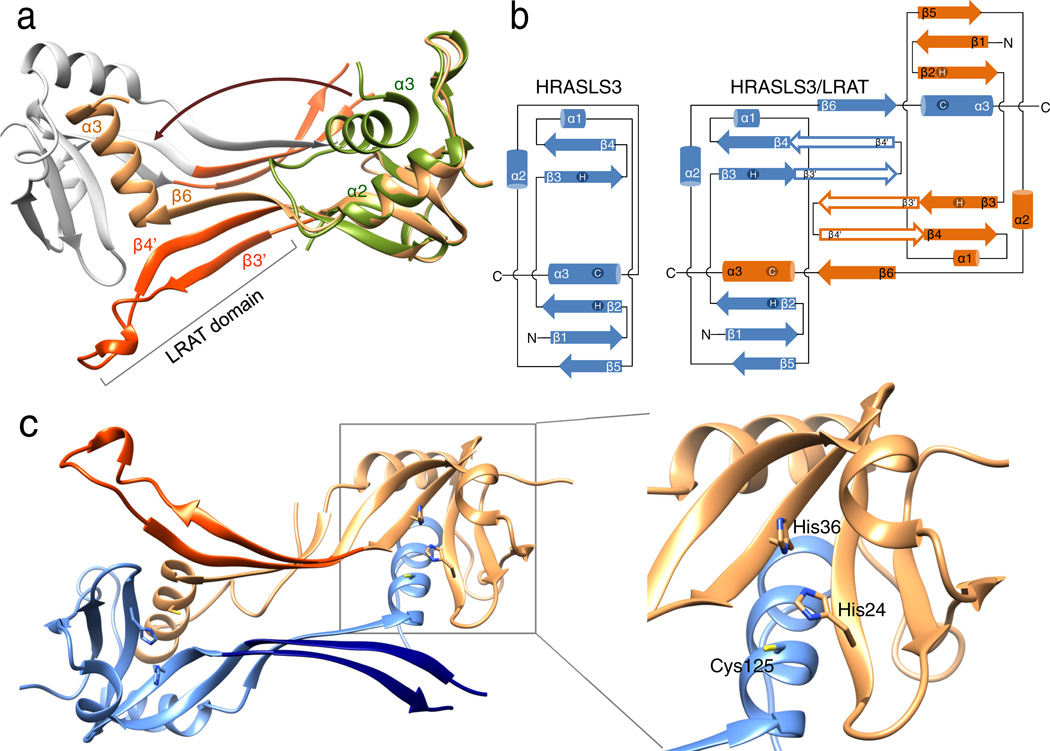
Gel filtration experiments suggested that the dominant form of the HRASLS3/LRAT chimera in solution is a dimer (Supplemental Fig. 8). Consequently, unlike the native HRASLS3 protein13,19,25, the chimeric enzyme crystallized in the form of a homo-dimer with local two-fold symmetry (Fig. 3a). The protein structure revealed two globular catalytic domains separated by two 3-stranded antiparallel β-sheets interacting with each other. An integral part of this linker was the 30-aa LRAT sequence that folds into a β-hairpin motif and constitutes the predominant dimerization interface reinforced by hydrophobic interactions between the side chains of the neighboring LRAT domain β-sheets (Fig. 3a; Supplementary Fig. 9). Interestingly, the dimerization process was accompanied by three-dimensional domain swapping in which the C-terminal α-helix 3 contributed to the structure of the adjacent protomer (Fig. 3a, b). This conformational rearrangement was enabled by the flexibility of a “hinge loop” between α-helix 2 and 3. Instead of turning around to complete the α/β fold of a monomer, it adopted an extended β-strand conformation (β6 in the domain-swapped dimer) and became part of the 3-stranded antiparallel β-sheet that constitutes the linker connecting the two catalytic domains. Stabilization of this newly adopted fold was possible due to extensive hydrogen bonding with the adjacent β-strand 4 that is part of the LRAT-domain β-hairpin fold. Notably, the “hinge loop” length was identical between HRASLS3 and LRAT indicating that native LRAT could easily be envisioned to adopt the same structure. Approximately 2415 Å2 of surface area was buried at the dimer interface for each of the monomers. The shape complementary value for the dimer interface was 0.72, indicating a highly complementary interaction comparable to other functionally dimeric enzymes26.
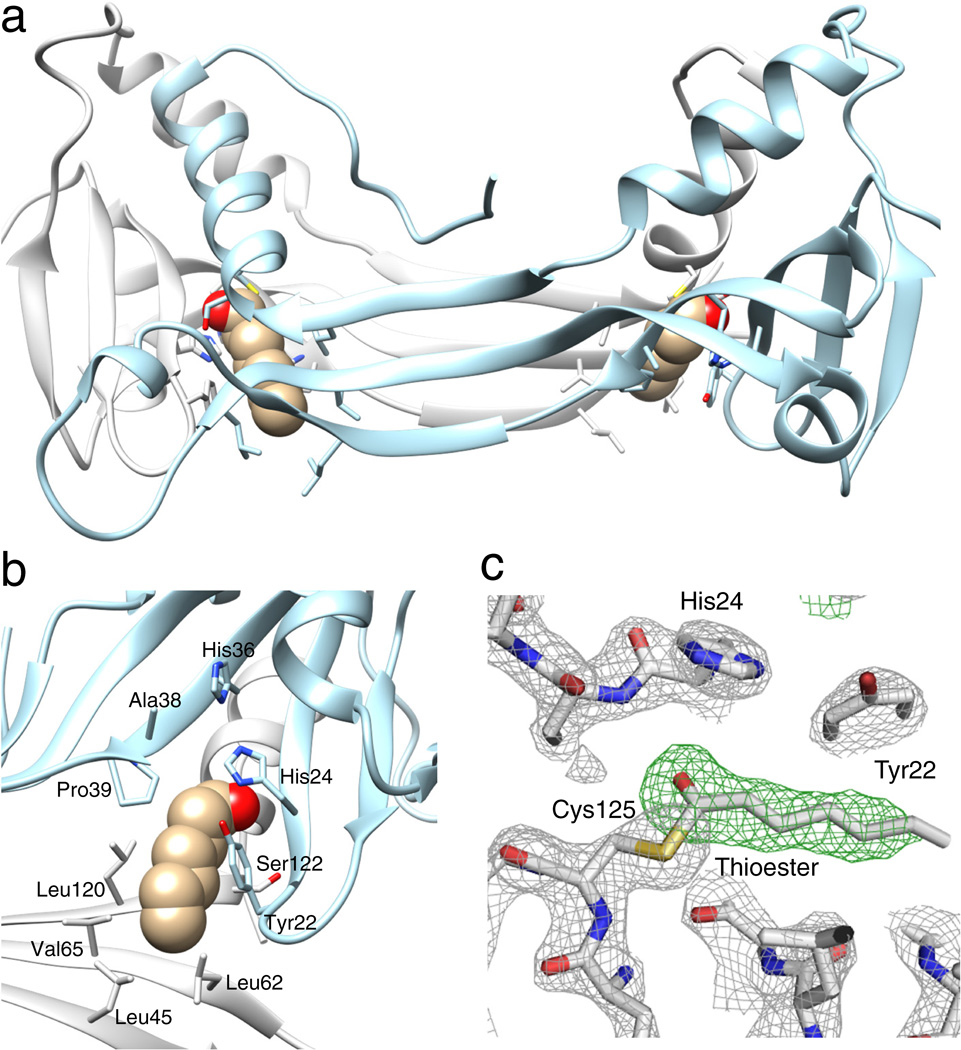
Figure 3. Crystal structure of the HRASLS3/LRAT chimeric protein.
(a) Ribbon diagram of HRASLS3/LRAT dimer with neighboring protomers colored in orange and gray. The LRAT-specific domain (red) adopts a β-hairpin structure. The superimposed structure of native HRASLS3 (PDB accession: 4DOT) is colored green. Dimerization-induced swapping of C-terminal α-helix 3 is marked with a brown arrow. (b) Protein topology diagrams for HRASLS3 and its chimeric counterpart. The scheme illustrates the protein chains’ locations in the intertwined dimeric structure and the role of the LRAT domains (β3’ and β4’) in providing the dimerization interface. (c) Configuration of the active sites within the HRASSL3/LRAT dimer. This protein structure reveals two globular catalytic domains linked together by two 3-stranded antiparallel β-sheets interacting with each other. The 30-aa LRAT sequences, colored in red and dark blue, are integral to the dimerization interface. Each active site is formed by residues donated by both members of the dimeric pair as shown in the zoom box.
Importantly, the swapped α-helix 3 contained the catalytic Cys125 residue. Thus, each of the two active sites present in the dimer is formed by residues donated from two neighboring polypeptide chains (Fig. 3c). Because the overall structures of the catalytic subunits within the domain-swapped oligomer are identical to that of the native HRASLS3, the active sites remained functional (Supplementary Fig. 10). However, the HRASLS3/LRAT chimera revealed important alterations of the active site environment. In contrast to HRASLS3 and HRASLS2 in which the catalytic residues are located in a shallow groove at the periphery of the protein13, the chimeric protein catalytic sites were embedded in a hydrophobic pocket lined by Leu45, 62, 120, Val65, Tyr22, and Pro39 side chains (Fig. 4a, b; Supplementary Fig. 10 and Supplementary Movie). Notably, the most hydrophobic Leu and Val residues were part of the 3-stranded antiparallel β-sheet formed by the LRAT domain of a neighboring protomer.
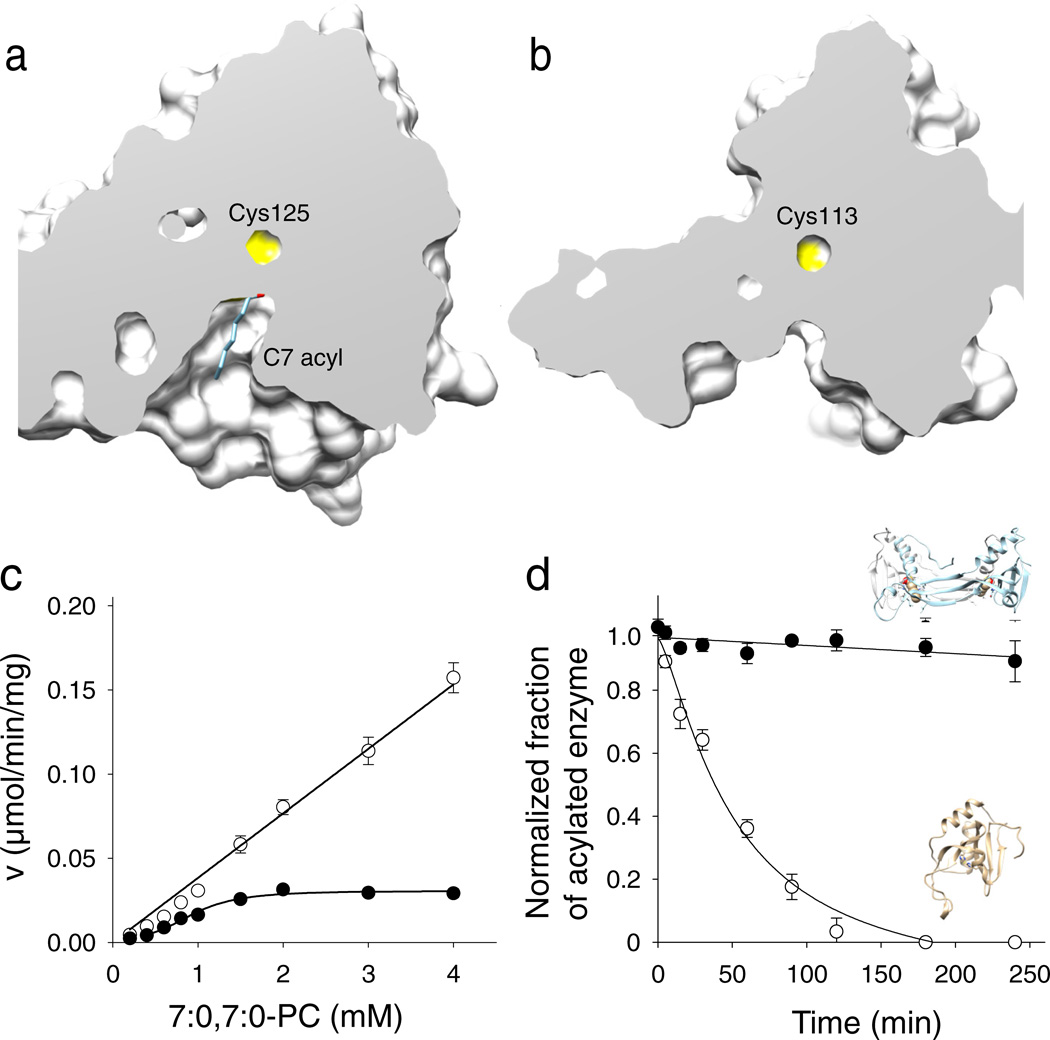
Figure 4. Effect of the structural rearrangement on the enzymatic actiivty.
Cut-away views of the protein surface at the active sites of HRASLS3/LRAT (a) and native HRASLS3 (b). Protein structures were aligned to ensure identical orientations. The NMR structure of HRASLS3 (PDB accession – 2KYT) was used for this figure. (c) Rates of phospholipid hydrolysis as a function of phospholipid substrate concentration for HRASLS3 (●) and its chimeric counterpart (○). (d) Stability of protein thioester adducts. The modified form of HRASLS3 steadily declines in the absence of phospholipid substrate, whereas the HRASLS3/LRAT thioester form remains intact for the duration of this experiment. Data represent mean values from three independent experiments performed in duplicates. Error bars, s.d.
Thus, the LRAT-specific sequence incorporated into HRASLS3 provides a basis for dimerization and a domain-swapping structural rearrangement by forming a major hydrophobic interaction interface between the monomers and by stabilizing the β-strand conformation of the hinge-loop region.
Structural changes affect the enzymatic activity
The structural rearrangements described above had important consequences for enzymatic function. These affected the active site environment, protein/lipid membrane interactions, and substrate binding allowing a new enzymatic activity for the chimeric protein. The buried position of the catalytic Cys125 in the chimera suggests that it is less accessible to water molecules and thus the thioester catalytic intermediate is more protected from hydrolysis than in the native HRASLS proteins. To test this hypothesis, we compared the rates of phospholipid cleavage and thioester intermediate hydrolysis in HRASLS3 and the corresponding chimeric enzyme after incubation with 7:0,7:0-PC. The enzymatic reaction velocity calculated for native HRASLS3 was much faster than that of the chimeric enzyme in each of the examined lipid concentrations (Fig. 4c). This difference in reaction rate was accompanied by an increased half-life of the thioester intermediate. LC/MS analyses of the intact proteins revealed that decay of the acyl-enzyme intermediate was much slower in the chimeric protein compared to its native counterpart (Fig. 4d, Supplementary Fig. 11). The stability of the acylated chimeric protein was sufficient to trap this reaction intermediate in the crystal form of the enzyme (Fig. 5a, b). The positive electron density, clearly seen in the Fo-Fc difference map corresponds to C7 acyl moiety donated by the lipid substrate added to the protein prior to crystallization (Fig. 5c). Despite extensive efforts, trapping acylated HRASLS3 in crystal form was not achieved suggesting that the greater solvent exposure of the HRASLS3 active site cavity promotes accelerated hydrolysis of the thioester bond.
Figure 5. The acylated form of HRASLS3/LRAT.
(a) Location of the acyl moieties within the HRASLS3/LRAT structure. (b) Organization of the hydrophobic pocket embedding the active site. The acyl moiety is represented with atomic spheres. (c) Electron density for the Cys125 acyl modification. Gray mesh represents a 2.2 Å resolution σA-weighted 2Fo − Fc electron density map contoured at 1.6σ. The green mesh corresponds to an unbiased σA-weighted Fo − Fc omit electron density map contoured at 3.5σ. The presence of the residual electron density extending from the Sγ atom of the Cys residue is highly suggestive of an acyl modification.
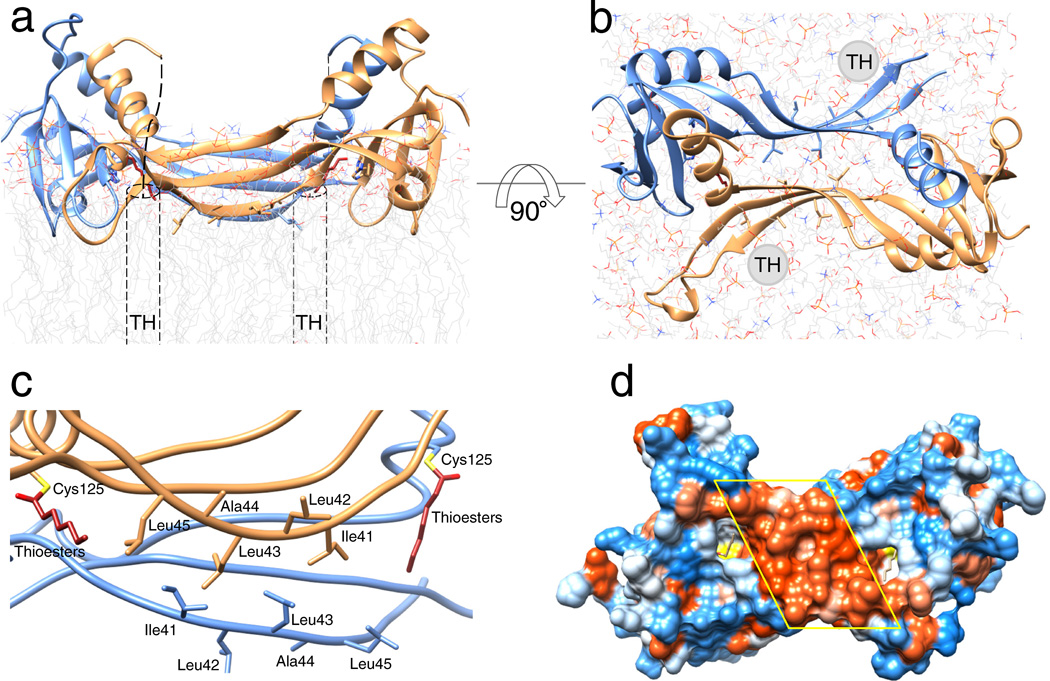
The well-defined all-trans conformation of the aliphatic carbon chain is determined by its interaction with hydrophobic residues that formed an envelope around the active site (Fig. 5; Supplementary Fig. 10 and 12). The overall size of this pocket was sufficient to accommodate an acyl chain of up to 8–10 carbons. However, the dominant acyl found at the sn-1 position of phospholipids was the palmitoyl moiety comprised of 16 carbon atoms. If the acyl chain length corresponded to that of the natural substrate, it would protrude well beyond the surface of the enzyme and its terminus would presumably remain embedded in the lipid membrane. Consequently, the membrane binding surface must position the hydrophobic active site groove in close proximity to the membrane interface to enable access to lipophilic, membrane-dissolved substrates. This surface thus defines the overall membrane topology of the protein (Fig. 6a, b). The interfacial surface includes β-strands formed by the N-terminal portion of the LRAT-specific sequence (40DILLALT46). In the dimeric structure, two of these β-strands belonging to the neighboring protomers were oriented anti-parallel to each other and formed a well-defined extended hydrophobic surface at the entrance to both active sites (Fig. 6c, d).
Figure 6. Phospholipid membrane topology of the HRASL3/LRAT chimera.
(a) and (b) Ribbon representations of the chimeric enzyme positioned at the lipid membrane viewed parallel and perpendicular to the membrane plane, respectively. Dashed cylinders represent putative positions of the C-terminal transmembrane α-helices (TM) absent in the crystallized protein. The overall topology was inferred from the parallel orientation of the acylated active sites and hydrophobicity of the protein/lipid interface. (c) Hydrophobic portion of the LRAT-specific domain with selected non-polar residues proposed to be involved in the lipid membrane interaction. (d) Hydrophobicity of the proposed membrane interaction surface. The molecular surface is colored according to the relative hydrophobicity of the side chains. Blue color corresponds to polar, whereas red indicates hydrophobic residues. The central hydrophobic patch corresponds to the membrane interacting region formed by residues indicated in panel c.
DISCUSSION
This biochemical and structural study of human N1pC/P60 proteins revealed an unexpected molecular adaptation that permits diversification of enzymatic specificity among this group of enzymes. Modification of the aa sequence outside of the catalytic core of HRASLS proteins appears to be crucial for adjustment of their enzymatic activity and ability to process vitamin A. The chimeric protein approach (Fig. 1 and 2) and examination of an LRAT deletion mutant (Supplementary Fig. 5) demonstrate that a 30-aa LRAT-specific sequence is necessary and sufficient for retinyl ester production but is not required for phospholipid hydrolysis.
Although structural and catalytic similarities between acyltransferases and hydrolases have been recognized, the molecular bases for their differing reactivities were not addressed. Hydrolysis is a common side reaction of acyltransferases, especially in the absence of a specific acceptor27,28. Thus, interactions between the acyl acceptor and the enzyme could contribute to this catalytic promiscuity. But comparison of the active site architecture of acyltransferases suggests that structural features such as orientation of the active site oxyanion loop influence the efficacy of water activation and thus favor acyl transfer over hydrolysis29. However, these mechanisms fail to explain the dramatic difference in phospholipid hydrolysis rates among native HRASLS enzymes and their chimeric counterparts in the absence of an acyl acceptor (Fig. 4c, d). Moreover, prolonged stability of the acylated form of the chimeric enzyme was not accompanied by structural changes within the putative oxyanion hole or the side chains that could affect activation of a nucleophilic water molecule (Supplementary Fig. 10 and 12). An alternative explanation for the altered activity could be limited access of water molecules to the active site. This hypothesis is supported by the structural comparison of HRASLS3/LRAT to native HRASLS3 (Fig. 4a, b; Supplementary Fig. 10). Due to the reduced size of the loop between β-strands 3 and 4 and its flexible conformations as documented by NMR, the catalytic Cys residue is fully accessible to solvent in HRASLS319. In contrast, the active site environment of the chimera is affected by a hydrophobic pocket that hosts the acyl moiety. The snug fit of the acyl chain further contributes to exclusion of water from the active site groove. LRAT is an integral membrane protein embedded in a phospholipid substrate. Given its constant exposure to acyl donors, this enzyme probably exists predominantly in the acylated form in native lipid membranes. We envision that interfacial protein binding provides an additional “seal” that in combination with the hydrophobic acyl binding channel further limits solvation of the active site. Thus, effective inhibition of thioester hydrolysis becomes the key adaptation that permits the utilization of acyl acceptors other than water or lyso-phospholipids.
Several lines of evidence indicate that the findings from the chimeric protein are directly relevant to native LRAT. The LRAT-specific sequence was necessary and sufficient to confer LRAT activity on the HRASLS proteins. This gain of function result together with the loss of LRAT activity (with retained PC hydrolase activity) when the LRAT-specific sequence was deleted from native LRAT demonstrates the importance of the LRAT-specific sequence in the enzymology of the protein. Moreover, dimerization and domain swapping found in the chimeric enzyme was induced by the LRAT-specific domain, which became an integral part of the dimerization interface. In fact, previous biochemical studies indicate that LRAT forms functional homodimers in microsomes isolated from the RPE as well as in purified and detergent solubilized forms30–32. Thus it is quite improbable that the same region could assume a radically different structure and function in native LRAT. Lastly, the aa sequence involved in the exclusion of water from the active site of the chimera is unique to LRAT and highly conserved among species indicating that the same molecular mechanism is applicable, regardless of the origin of the enzyme.
Replacement of a sequence between the β3 and β4 strands in the catalytic domain induced a structural rearrangement that formed a 3D domain-swapped dimer with two independent catalytic domains (Fig. 3c). Notably, hydrophobic shielding of the active site of one protomer was accomplished in large part by residues contributed by its dimeric partner. Thus, dimerization appears to be indispensable for LRAT enzymatic activity. A functional role of dimerization would therefore explain the associated C-terminal domain swapping. In addition to the interaction between the LRAT domains, the swapped α-helix 3 and the “hinge” region further contributed to the intermolecular interface (Fig. 3a). This increased area for monomer interactions reinforces the stability of the dimer. Thus, the HRASLS3/LRAT chimera and, by inference, LRAT itself are examples of enzymes that depend on 3D domain swapping for their biological function.
Divergence of enzymes via gene duplication, mutation, and selection has led to the present diversity of enzymatic activities and metabolic competence33. The efficiency of this process depends on the ability of nature to modify preexisting common structural and functional features to adopt new roles34–36. An iconic example of this process is the evolution of enzymes constituting the visual cycle, a metabolic pathway that regenerates visual pigment in jawed vertebrates37–39. Rather than by an earlier evolutionary photoisomerization, the key activity that enables enzymatic regeneration of 11-cis-retinal (visual chromophore) is esterification of vitamin A (all-trans-retinol) via the action of LRAT, the obligatory source for all-trans-retinyl esters for retinoid isomerase (RPE65)7,40,41. The basis for the interfacial enzymatic activity of LRAT can now be inferred from the structure of the HRASLS3/LRAT chimeric protein. Although the examined protein lacks a C-terminal transmembrane helix, the location of trapped acyl moieties in close proximity to a hydrophobic patch on the protein surface provides sufficient information to determine a preferential orientation of these molecules with respect to the phospholipid membrane. Because docking to the active site at the phospholipid bilayer allows substrate accessibility without a thermodynamically unfavorable extraction of lipids from the membrane, we assume that the interfacial binding surface is adjacent to the mouth of the hydrophobic channel (Fig. 6). Although, the primary structural element that allows membrane insertion is generally a C-terminal transmembrane helix, secondary protein-lipid interactions are probably critical for proper active site membrane embedding and could affect substrate specificity. The dominant region involved in lipid interactions is the enlarged hydrophobic patch formed by two Leu- and Ile-rich fragments of the LRAT-specific sequence that belong to its neighboring monomers (Fig. 6c, d). These symmetric and highly hydrophobic β-strands oriented parallel to the lipid-water interface represent an unusual mode of protein-lipid interaction. This hydrophobic plateau domain allows a monotopic mode of interaction that reinforces bilayer binding and possibly allows both catalytic subunits to function concurrently by recruiting substrate from the same membrane. This dominant hydrophobic patch is most likely responsible for interaction of tLRAT with artificial lipid membranes documented by physicochemical methods42.
Although the structure of HRASLS3/LRAT in its acylated form is presumably the most competent to interact with retinoids, it does not provide direct evidence for a vitamin A binding site. Retinoids are relatively large and rigid ligands that typically occupy well-defined binding sites43,44. Lack of such an obvious feature in the chimeric protein indicates that, similar to the phospholipid substrate, all-trans-retinol is obtained directly from the lipid membrane and attracted to the active site by hydrophobic interactions with a portion of the LRAT-specific domain. This model could explain the broad LRAT specificity for retinols, including their geometric isomers as well as their derivatives with a modified β-ionone ring and polyene chain45.
In summary, biochemical and structural analyses of vertebrate representatives of the NlpC/P60 protein family offer valuable insights into the molecular adaptations that lead to acquisition of novel substrate specificity by modifying existing structural motifs. We identified a key sequence modification that determines diversification of enzymatic activities within a closely related enzyme family and describe its role at a molecular level. We also provide evidence for the functional significance of 3D-domain swapping dimerization in ensuring a proper active site environment.
ONLINE METHODS
Materials
cDNAs encoding human HRASLS2, and HRASLS4 (gi:8923526, and gi:149588791) as well as mouse LRAT (gi:12963753, sequence encoding residues 30–186, tLRAT), its deletion mutant (Δ76–81/Δ90–102) and HRASLS2, −3, and −4/LRAT chimeric proteins were chemically synthesized de novo (DNA 2.0). A clone of human HRASLS3 (gi:189571621) was received from the American Type Culture Collection. Phospholipids were purchased from Avanti Polar Lipids. Protein crystallization screens were obtained from Hampton Research. All reagents were of the highest purity commercially available.
Expression and purification of LRAT, HRASLS, and HRASLS/LRAT chimeric proteins
Expression and purification of soluble forms of HRASLS2 (residues 1–129), HRASLS3 (residues 1–132), and HRASLS4 (residues 1–134) enzymes were carried out as described in our previous report13.
To obtain functional LRAT and its deletion mutant, mouse cDNA was subcloned into a pGEX_2T vector (GE Healthcare) using BamHI and EcoRI restriction sites to generate an N-terminal in frame fusion with the sequence encoding glutathione S-transferase. The expressed fusion protein (GST-tLRAT) was purified by the procedure described previously4.
HRASLS/LRAT chimeric proteins were constructed by replacing the native sequence of HRASLS2, 3, and 4 (residues 39 – 57) with the 30-aa mouse LRAT sequence (76DILLALTNDKERTQKVVSNKRLLLGVICKV106) (Fig. 1). These chimeric enzymes were expressed as glutathione S-transferase (GST) fusion proteins. cDNAs of HRASLS2, 3, and 4/LRAT were subcloned into a pGEX_2T vector using BamHI and EcoRI restriction sites and expressed in the BL21(DE3) E. coli strain. Bacteria were grown in a shaker incubator in the presence of 50 µm ampicillin and protein expression was induced with 0.5 mM isopropyl β-D-1-thiogalactopyranoside. After 5 h at 30 °C, bacteria were harvested and disrupted by osmotic shock46. The crude bacterial lysate was centrifuged at 36,000g at 4 °C for 30 min. The buffer composition of the soluble fraction was adjusted to pH 7.4 with 67 mM phosphate buffer and 50 mM NaCl. Then the extract was incubated with glutathione-Sepharose resin (GE Healthcare) for 2 h at 4 °C. The resin was placed in a chromatography column and washed with 10 column volumes of 67 mM phosphate buffer, pH 7.4, and 50 mM NaCl. GST-HRASLS/LRAT fusion chimeric proteins were eluted with 10 mM reduced glutathione in 67 mM phosphate buffer, pH 7.4, and 50 mM NaCl. Protein-containing fractions were pooled and concentrated in a 30 kDa cutoff Centricon (Amicon) to a final volume of 5 mL. The N-terminal GST tag was removed by digestion of the fusion protein with thrombin (2 activity units per 1 mg of protein) at room temperature for 4 h with completion of the digestion monitored by SDS-PAGE. The protein solution was then diluted 10 times with 10 mM MES/NaOH buffer, pH 6.2, 5 mM DTT and loaded onto a HiTrap SP-sepharose column (GE Healthcare). At pH 6.2, HRASLS/LRAT chimeric proteins bound preferentially to the resin whereas the cleaved GST appeared in flow-through fractions. Protein trapped on the SP-column was eluted with a linear gradient of NaCl (0 – 0.5 M) in 10 mM MES/NaOH buffer, pH 6.2, 5 mM DTT. The purity of fractionated proteins was examined by SDS-PAGE (Fig. S3). Fractions containing virtually pure proteins were pooled together, concentrated to ~2 mg/mL in a 10 kDa cutoff Centricon, snap frozen in liquid nitrogen, and stored at −80 °C. The molecular mass and aa sequence of purified chimeric proteins were confirmed by mass spectrometry13.
Retinol acyltransferase enzymatic activity assay
Formation of retinyl esters was assayed in 0.2 mL of 10 mM Tris/HCl buffer, pH 8.0, 5 mm DTT, 1% BSA (w/w), and 5 µg of purified proteins at 37 °C. The reaction was initiated by addition of 2 mM 1,2-diheptanoyl-sn-glycero-3-phosphocholine (7:0,7:0-PC) along with all-trans-retinol (Toronto Research Chemicals) delivered in 1 µL of dimethylformamide to a final concentration of 20 µM. The reaction mixture was vigorously vortexed, incubated at 37 °C for 1 h, and then quenched with 0.2 mL of methanol followed by retinoid extraction with 0.3 mL of hexane. The composition of organic extracts was analyzed by HPLC on a normal phase silica column (Agilent Sil 5 µm, 4.6 × 250 mm) (Agilent Technology). Retinoids were separated in a stepwise gradient of 2% ethyl acetate in hexane for the initial 10 min and then 20% ethyl acetate for an additional 20 min at a flow rate of 1.4 mL/min. Retinyl ester products were detected, identified, and quantified based on their UV-visible absorbance and comparison of their retention times to those of synthetic standards. For kinetic studies, the initial rates of all-trans-retinol esterification were measured at retinoid substrate concentrations ranging between 2 to 200 µM. Incubation times were adjusted for each of the tested enzymes according to their reaction rates (5 min for LRAT, 15 min for HRASLS2/LRAT and HRASLS4/LRAT, and 60 min for HRASLS3/LRAT). Kinetic parameters were calculated based on the nonlinear regression of the experimental data fitted with the Michaelis-Menten model.
All-trans-retinol fluorescence binding assay
Binding of all-trans-retinol to HRASLS and HRASLS/LRAT chimeric proteins was studied by spectrofluorometry47. Measurements were performed with a PerkinElmer Life Sciences LS55 model fluorometer (Watman). Interaction of all-trans-retinol with proteins was assessed by monitoring the quenching of protein fluorescence with increasing concentrations of ligand. Samples were excited at 285 nm. Emission spectra were recorded between 300 and 520 nm with bandwidths for excitation and emission set to 10 nm. All titrations were carried out at 25 °C in 20 mM Tris/HCl buffer, pH 8.0, containing 50 mM NaCl and 10% glycerol (v/v). all-trans-Retinol was delivered in methanol such that the final volume of this organic solvent did not exceed 0.5% of the sample's total volume. All binding data were corrected for background and self-absorption of excitation and emission light48. Apparent Kd values were calculated based on nonlinear regression of the experimental data and using one or two site saturation ligand-binding models.
Phosphatidylcholine cleavage and transesterification assay
7:0,7:0-PC or 1-hexanoyl-2-(6-((7-nitro-2-1,3-benzoxadiazol-4-yl)amino)hexanoyl)-sn-glycero-3-phosphocholine (NBD-PC) at concentrations of 2.0 and 0.1 mM, respectively, was incubated with 5 µg of purified enzymes in 10 mM Tris/HCl buffer, pH 8.0, 1 mM DTT, 5% methanol for 30 min (native HRASLS proteins) or 3 h (chimeras, GST-tLRAT, and Δ76–81/Δ90–102 LRAT deletion mutant) at 30 °C. The lipid composition of the reaction mixture was directly examined by LC/MS. An Agilent Technologies 1100 Series HPLC system equipped with a diode array UV/Vis detector was interfaced with an LXQ mass spectrometer (Thermo Scientific). Substrate and products of the reaction were separated on an XBridge BEH300 C4 column (3.5 µm, 2.1 × 50 mm) (Waters) with a linear (5–100%) gradient of acetonitrile in water developed over 20 min at a flow rate of 0.25 mL/min; both solvents contained 0.1% formic acid. The eluate was first directed into the UV/Vis detector followed by the electrospray ionization probe and mass spectrometer. Products of the enzymatic reaction were quantified based on their absorbance at 460 nm whereas the chemical identities of eluted compounds were confirmed by MS as previously described13.
Self-acylation and thioester intermediate hydrolysis assays
Acylated forms of HRASLS3 and its LRAT chimera were readily detected in the presence of short acyl chain PCs by recording their intact protein MS spectra13. One-half a µg of protein was loaded onto an XBridge BEH300 C4 column (3.5 µm, 2.1 × 50 mm) and eluted with a linear gradient (5 – 100%) of acetonitrile in water as described above. Intact protein masses were deconvoluted manually or by using the ProMass module for Xcalibur software (Thermo Scientific). To determine the apparent stability of the thioester form, enzymes were pre-incubated with 1 mM 7:0,7:0-PC in 10 mM MES-NaOH, pH 6.3, 0.2 M NaCl and 1 mM DTT for 10 min at 25 °C. Then excess PC was removed by loading the protein solution onto a PD-10 desalting column (GE Healthcare) equilibrated with 10 mM Tris/HCl buffer, pH 8.0, 1 mM DTT. Eluted proteins were incubated for various time periods at 25 °C and subsequently snap frozen and stored in liquid nitrogen. Samples were thawed, one by one, directly prior to MS analysis. The fraction of enzymes that remained acylated was calculated based on the ratio between intact protein intensity ions of unmodified and acylated forms of each enzyme.
Crystallization, X-ray data collection and processing
Crystals of the HRASLS3/LRAT chimeric protein were grown by the sitting drop vapor diffusion method. Prior to crystallization, the protein solution (1.5 mg/ml) in 10 mM MES-NaOH, pH 6.3, containing 0.2 M NaCl and 10 mM DTT was incubated with 10 mM 7:0,7:0-PC for 2 min on ice. Crystallization was initiated by mixing 2 µL of the protein solution with 2 µL of crystallization solution consisting of 0.1 M Tris/HCl, pH 8.5, 0.2 M NaCl, and 20% (w/v) polyethylene glycol 3350. The resulting drops were incubated at room temperature over 0.5 mL of the same crystallization solution. Crystals were typically observed after 10 to 16 h. Most crystals were tetragonal and reached dimensions of 25 to 50 µm. Freshly grown crystals were harvested and directly flash cooled in liquid nitrogen prior to X-ray exposure.
A high resolution data set extending to 2.2 Å resolution was collected at the Advanced Photon Source NE-CAT 24-ID-C beamline. Data were processed by using XDS49 and programs from the CCCP4 suite50. Crystals belonged to space group P43212 and contained two monomers per asymmetric unit (supplemental Table 2). The structure of HRASLS3/LRAT was solved by molecular replacement with PHASER_MR51 using the atomic coordinates of HRASLS3 (Protein Data Bank accession code: 4DOT) as a search model. Model refinement was carried out with REFMAC52 and completed and manually adjusted with COOT53. The residual positive electron density that emanated from the active site Cys sulfhydryl was modeled as a covalent heptanoyl thioester group based on its appearance and the surrounding chemical environment. External distance and planarity restraints were applied to the thioester moiety during refinement with REFMAC. TLS modeling of the atomic displacement parameters was applied in the later stages of refinement with groups determined by the TLSMD web server54. The structure refinement converged to an Rfree value of 24.76% with 97% favored and no Ramachandran plot outliers as assessed with the Molprobity server55. Residues 1 – 6 in chain A and 1 – 4, 51 – 59, and 141 – 147 in chain B were omitted from the final model because of weak or absent electron densities. Fifty eight water molecules were included in the final model.
Statistical analysis
Data representing the means ± s.d. for the results of at least three independent experiments were compared by the one-way analysis of variance test.
Supplementary Material
Acknowledgments
We thank Drs. Xiongying Tu and Brian M. Kevany for initial X-ray diffraction data collection of HRASLS3/LRAT crystals, and Dr. Leslie T. Webster, Jr. for help in preparation of this manuscript. This work was supported by grants EY023948 (M.G.), Nutrition and Obesity Research Center (M.G.), and EY009339 (K.P.) from the National Eye Institute of the National Institutes of Health (NIH) as well as MSTP training grant, T32 GM007250 (A.E.S.). We thank the NE-CAT staff for assistance with diffraction data collection. Use of the Advanced Photon Source, an Office of the Science User Facility operated for the United States Department of Energy Office of Science by the Argonne National Laboratory, was supported by the United States Department of Energy under Contract DE-AC02-06CH11357. Preliminary data for this study were obtained at beamline X29 of the National Synchrotron Light Source. Its financial support is derived principally from the Offices of Biological and Environmental Research and of Basic Energy Sciences of the United States Department of Energy and by National Institutes of Health Grant P41RR012408 from NCRR and P41GM103473 from NIGMS. KP is John H. Hord Professor of Pharmacology.
Abbreviations
- aa
amino acid
- GST
glutathione S-transferase
- HPLC
high performance liquid chromatography
- HRASLS
H-Ras-like tumor suppressors
- LC/MS
liquid chromatography-mass spectrometry
- LRAT
lecitihin:retinol acyltransferase
- MS
mass spectrometry
- NBD-PC
1-hexanoyl-2-(6-((7-nitro-2-1,3-benzoxadiazol-4-yl)amino)hexanoyl)-sn-glycero-3-phosphocholine
- PC
phosphatidylcholine
- PE
phosphatidylethanolamine
- RPE
retinal pigment epithelium
- RPE65
retinal pigment epithelium-specific 65 kDa protein
- tLRAT
truncated mouse lecitihin:retinol acyltransferase, sequence encoding residues 30 – 186
Footnotes
Contributions
M.G. and K. P. designed the experiments. M.G. and A.E.S. performed biochemical experiments and crystalized HRASLS3/LRAT. M.G. and P.D.K. collected and processed the crystallographic data. All authors contributed to the data analyses. M.G., A.E.S. wrote the manuscript with valuable input from P.D.K. and K.P.
Accession codes
The coordinates and structure factor amplitudes were deposited in the Protein Data Bank under accession code 4Q95.
Declaration of competing financial interest
We declare that the authors have no competing interests as defined by Nature Publishing Group, or other interests that might be perceived to influence the results and discussion reported in this paper.
REFERENCES
- 1.Wymann MP, Schneiter R. Lipid signalling in disease. Nat Rev Mol Cell Biol. 2008;9:162–176. doi: 10.1038/nrm2335. [DOI] [PubMed] [Google Scholar]
- 2.Forneris F, Mattevi A. Enzymes without borders: mobilizing substrates, delivering products. Science. 2008;321:213–216. doi: 10.1126/science.1151118. [DOI] [PubMed] [Google Scholar]
- 3.Derewenda ZS. Structure and function of lipases. Adv Protein Chem. 1994;45:1–52. doi: 10.1016/s0065-3233(08)60637-3. [DOI] [PubMed] [Google Scholar]
- 4.Golczak M, Palczewski K. An acyl-covalent enzyme intermediate of lecithin:retinol acyltransferase. J Biol Chem. 2010;285:29217–29222. doi: 10.1074/jbc.M110.152314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anantharaman V, Aravind L. Evolutionary history, structural features and biochemical diversity of the NlpC/P60 superfamily of enzymes. Genome Biol. 2003;4:R11. doi: 10.1186/gb-2003-4-2-r11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Albalat R. Evolution of the genetic machinery of the visual cycle: a novelty of the vertebrate eye? Mol Biol Evol. 2012;29:1461–1469. doi: 10.1093/molbev/msr313. [DOI] [PubMed] [Google Scholar]
- 7.Batten ML, et al. Lecithin-retinol acyltransferase is essential for accumulation of all-trans-retinyl esters in the eye and in the liver. Journal of Biological Chemistry. 2004;279:10422–10432. doi: 10.1074/jbc.M312410200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu LM, Gudas LJ. Disruption of the lecithin: Retinol acyltransferase gene makes mice more susceptible to vitamin a deficiency. Journal of Biological Chemistry. 2005;280:40226–40234. doi: 10.1074/jbc.M509643200. [DOI] [PubMed] [Google Scholar]
- 9.Amengual J, Golczak M, Palczewski K, von Lintig J. Lecithin: Retinol Acyltransferase Is Critical for Cellular Uptake of Vitamin A from Serum Retinol-binding Protein. Journal of Biological Chemistry. 2012;287:24216–24227. doi: 10.1074/jbc.M112.353979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Borman AD, et al. Early Onset Retinal Dystrophy Due to Mutations in LRAT: Molecular Analysis and Detailed Phenotypic Study. Invest Ophthalmol Vis Sci. 2012;53:3927–3938. doi: 10.1167/iovs.12-9548. [DOI] [PubMed] [Google Scholar]
- 11.Jaworski K, et al. AdPLA ablation increases lipolysis and prevents obesity induced by high-fat feeding or leptin deficiency. Nat Med. 2009;15:159–168. doi: 10.1038/nm.1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wolf G. Adipose-specific phospholipase as regulator of adiposity. Nutr Rev. 2009;67:551–554. doi: 10.1111/j.1753-4887.2009.00227.x. [DOI] [PubMed] [Google Scholar]
- 13.Golczak M, et al. Structural Basis for the Acyltransferase Activity of Lecithin:Retinol Acyltransferase-like Proteins. Journal of Biological Chemistry. 2012;287:23790–23807. doi: 10.1074/jbc.M112.361550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Uyama T, Jin XH, Tsuboi K, Tonai T, Ueda N. Characterization of the human tumor suppressors TIG3 and HRASLS2 as phospholipid-metabolizing enzymes. Biochim Biophys Acta. 2009;1791:1114–1124. doi: 10.1016/j.bbalip.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 15.Uyama T, et al. Generation of N-acylphosphatidylethanolamine by members of the phospholipase A/acyltransferase (PLA/AT) family. J Biol Chem. 2012;287:31905–31919. doi: 10.1074/jbc.M112.368712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Han BG, et al. Expression, purification and biochemical characterization of the N-terminal regions of human TIG3 and HRASLS3 proteins. Protein Expr Purif. 2010;71:103–107. doi: 10.1016/j.pep.2010.01.018. [DOI] [PubMed] [Google Scholar]
- 17.MacDonald PN, Ong DE. A lecithin:retinol acyltransferase activity in human and rat liver. Biochem Biophys Res Commun. 1988;156:157–163. doi: 10.1016/s0006-291x(88)80818-0. [DOI] [PubMed] [Google Scholar]
- 18.Jin XH, et al. Discovery and characterization of a Ca2+-independent phosphatidylethanolamine N-acyltransferase generating the anandamide precursor and its congeners. J Biol Chem. 2007;282:3614–3623. doi: 10.1074/jbc.M606369200. [DOI] [PubMed] [Google Scholar]
- 19.Ren X, Lin J, Jin C, Xia B. Solution structure of the N-terminal catalytic domain of human H-REV107--a novel circular permutated NlpC/P60 domain. FEBS Lett. 2010;584:4222–4226. doi: 10.1016/j.febslet.2010.09.015. [DOI] [PubMed] [Google Scholar]
- 20.Wang L, Cvetkov TL, Chance MR, Moiseenkova-Bell VY. Identification of in vivo disulfide conformation of TRPA1 ion channel. J Biol Chem. 2012;287:6169–6176. doi: 10.1074/jbc.M111.329748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saari JC, Bredberg DL. Lecithin:retinol acyltransferase in retinal pigment epithelial microsomes. J Biol Chem. 1989;264:8636–8640. [PubMed] [Google Scholar]
- 22.Bolen AL, et al. The phospholipase A1 activity of lysophospholipase A-I links platelet activation to LPA production during blood coagulation. J Lipid Res. 2011;52:958–970. doi: 10.1194/jlr.M013326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sano T, et al. Multiple mechanisms linked to platelet activation result in lysophosphatidic acid and sphingosine 1-phosphate generation in blood. J Biol Chem. 2002;277:21197–21206. doi: 10.1074/jbc.M201289200. [DOI] [PubMed] [Google Scholar]
- 24.Uyama T, et al. The tumor suppressor gene H-Rev107 functions as a novel Ca2+-independent cytosolic phospholipase A1/2 of the thiol hydrolase type. J Lipid Res. 2009;50:685–693. doi: 10.1194/jlr.M800453-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pang XY, et al. Structure/function relationships of adipose phospholipase A2 containing a cys-his-his catalytic triad. J Biol Chem. 2012;287:35260–35274. doi: 10.1074/jbc.M112.398859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lawrence MC, Colman PM. Shape complementarity at protein/protein interfaces. J Mol Biol. 1993;234:946–950. doi: 10.1006/jmbi.1993.1648. [DOI] [PubMed] [Google Scholar]
- 27.Liu M, Subbaiah PV. Hydrolysis and transesterification of platelet-activating factor by lecithin-cholesterol acyltransferase. Proc Natl Acad Sci U S A. 1994;91:6035–6039. doi: 10.1073/pnas.91.13.6035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fournand D, Arnaud A. Aliphatic and enantioselective amidases: from hydrolysis to acyl transfer activity. J Appl Microbiol. 2001;91:381–393. doi: 10.1046/j.1365-2672.2001.01378.x. [DOI] [PubMed] [Google Scholar]
- 29.Jiang Y, Morley KL, Schrag JD, Kazlauskas RJ. Different active-site loop orientation in serine hydrolases versus acyltransferases. Chembiochem. 2011;12:768–776. doi: 10.1002/cbic.201000693. [DOI] [PubMed] [Google Scholar]
- 30.Jahng WJ, Cheung E, Rando RR. Lecithin retinol acyltransferase forms functional homodimers. Biochemistry. 2002;41:6311–6319. doi: 10.1021/bi015710b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bok D, et al. Purification and characterization of a transmembrane domain-deleted form of lecithin retinol acyltransferase. Biochemistry. 2003;42:6090–6098. doi: 10.1021/bi0342416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Golczak M, Kiser PD, Lodowski DT, Maeda A, Palczewski K. Importance of membrane structural integrity for RPE65 retinoid isomerization activity. J Biol Chem. 2010;285:9667–9682. doi: 10.1074/jbc.M109.063941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Conant GC, Wolfe KH. Turning a hobby into a job: how duplicated genes find new functions. Nat Rev Genet. 2008;9:938–950. doi: 10.1038/nrg2482. [DOI] [PubMed] [Google Scholar]
- 34.Khersonsky O, Tawfik DS. Enzyme promiscuity: a mechanistic and evolutionary perspective. Annu Rev Biochem. 2010;79:471–505. doi: 10.1146/annurev-biochem-030409-143718. [DOI] [PubMed] [Google Scholar]
- 35.O'Brien PJ, Herschlag D. Catalytic promiscuity and the evolution of new enzymatic activities. Chem Biol. 1999;6:R91–R105. doi: 10.1016/S1074-5521(99)80033-7. [DOI] [PubMed] [Google Scholar]
- 36.Jacob F. Evolution and tinkering. Science. 1977;196:1161–1166. doi: 10.1126/science.860134. [DOI] [PubMed] [Google Scholar]
- 37.Albalat R. Evolution of the Genetic Machinery of the Visual Cycle: A Novelty of the Vertebrate Eye? Mol Biol Evol. 2012;29:1461–1469. doi: 10.1093/molbev/msr313. [DOI] [PubMed] [Google Scholar]
- 38.Kusakabe TG, Takimoto N, Jin MH, Tsuda M. Evolution and the origin of the visual retinoid cycle in vertebrates. Philosophical Transactions of the Royal Society B-Biological Sciences. 2009;364:2897–2910. doi: 10.1098/rstb.2009.0043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Poliakov E, et al. Origin and Evolution of Retinoid Isomerization Machinery in Vertebrate Visual Cycle: Hint from Jawless Vertebrates. PLoS One. 2012;7 doi: 10.1371/journal.pone.0049975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jin MH, Li SH, Moghrabi WN, Sun H, Travis GH. Rpe65 is the retinoid isomerase in bovine retinal pigment epithelium. Cell. 2005;122:449–459. doi: 10.1016/j.cell.2005.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Redmond TM, et al. Mutation of key residues of RPE65 abolishes its enzymatic role as isomerohydrolase in the visual cycle. Proc Natl Acad Sci U S A. 2005;102:13658–13663. doi: 10.1073/pnas.0504167102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bussieres S, Cantin L, Desbat B, Salesse C. Binding of a truncated form of lecithin:retinol acyltransferase and its N- and C-terminal peptides to lipid monolayers. Langmuir. 2012;28:3516–3523. doi: 10.1021/la203896n. [DOI] [PubMed] [Google Scholar]
- 43.Kiser PD, Golczak M, Palczewski K. Chemistry of the retinoid (visual) cycle. Chem Rev. 2014;114:194–232. doi: 10.1021/cr400107q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pakhomova S, Kobayashi M, Buck J, Newcomer ME. A helical lid converts a sulfotransferase to a dehydratase. Nat Struct Biol. 2001;8:447–451. doi: 10.1038/87617. [DOI] [PubMed] [Google Scholar]
- 45.Canada FJ, et al. Substrate specificities and mechanism in the enzymatic processing of vitamin A into 11-cis-retinol. Biochemistry. 1990;29:9690–9697. doi: 10.1021/bi00493a026. [DOI] [PubMed] [Google Scholar]
- 46.Burger A, Berendes R, Voges D, Huber R, Demange P. A rapid and efficient purification method for recombinant annexin V for biophysical studies. FEBS Lett. 1993;329:25–28. doi: 10.1016/0014-5793(93)80185-w. [DOI] [PubMed] [Google Scholar]
- 47.Golczak M, et al. Metabolic basis of visual cycle inhibition by retinoid and nonretinoid compounds in the vertebrate retina. J Biol Chem. 2008;283:9543–9554. doi: 10.1074/jbc.M708982200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lakowicz JR. Principles of fluorescence spectroscopy. xxvi. New York: Springer; 2006. p. 954. [Google Scholar]
- 49.Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mccoy AJ, et al. Phaser crystallographic software. Journal of Applied Crystallography. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallographica Section D-Biological Crystallography. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 53.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallographica Section D-Biological Crystallography. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 54.Painter J, Merritt EA. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr D Biol Crystallogr. 2006;62:439–450. doi: 10.1107/S0907444906005270. [DOI] [PubMed] [Google Scholar]
- 55.Davis IW, et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.