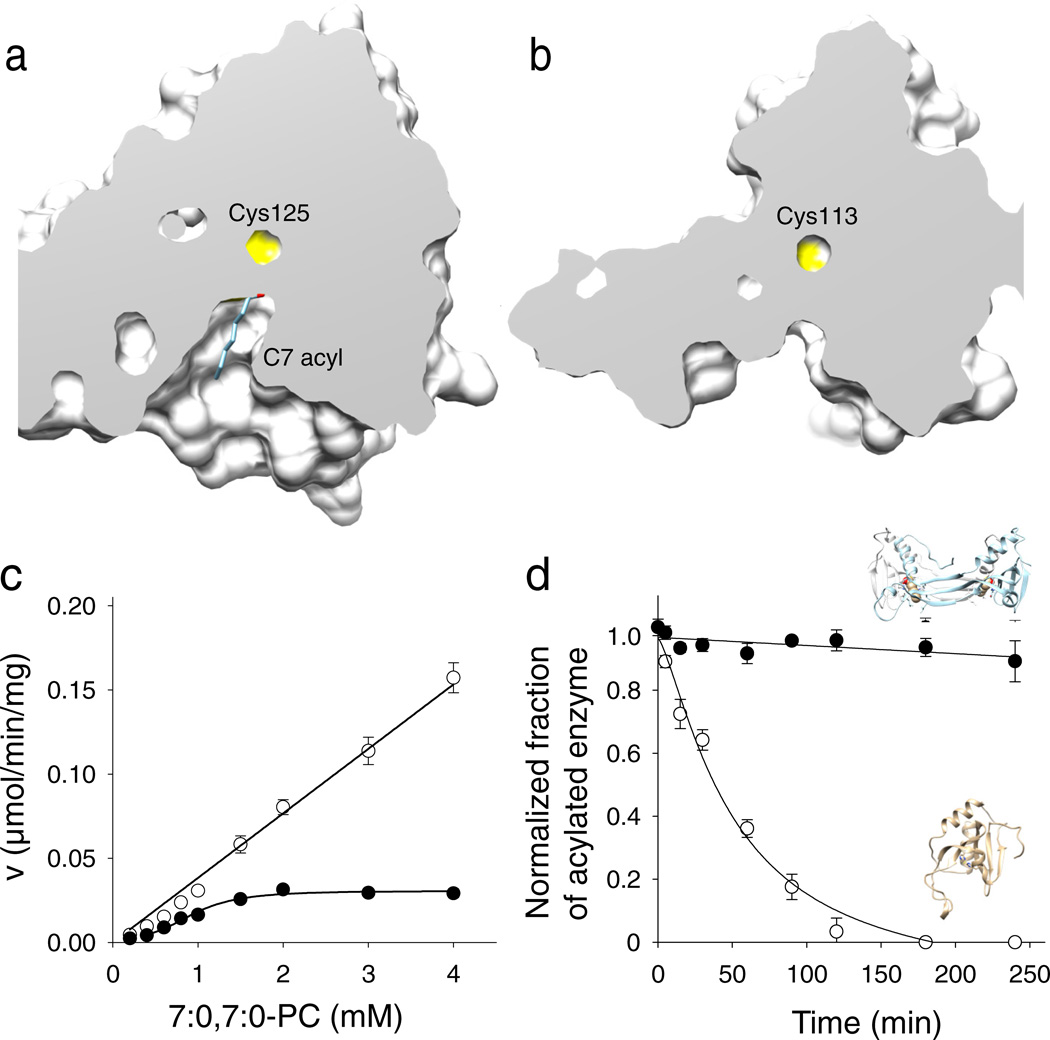
Figure 4. Effect of the structural rearrangement on the enzymatic actiivty.
Cut-away views of the protein surface at the active sites of HRASLS3/LRAT (a) and native HRASLS3 (b). Protein structures were aligned to ensure identical orientations. The NMR structure of HRASLS3 (PDB accession – 2KYT) was used for this figure. (c) Rates of phospholipid hydrolysis as a function of phospholipid substrate concentration for HRASLS3 (●) and its chimeric counterpart (○). (d) Stability of protein thioester adducts. The modified form of HRASLS3 steadily declines in the absence of phospholipid substrate, whereas the HRASLS3/LRAT thioester form remains intact for the duration of this experiment. Data represent mean values from three independent experiments performed in duplicates. Error bars, s.d.