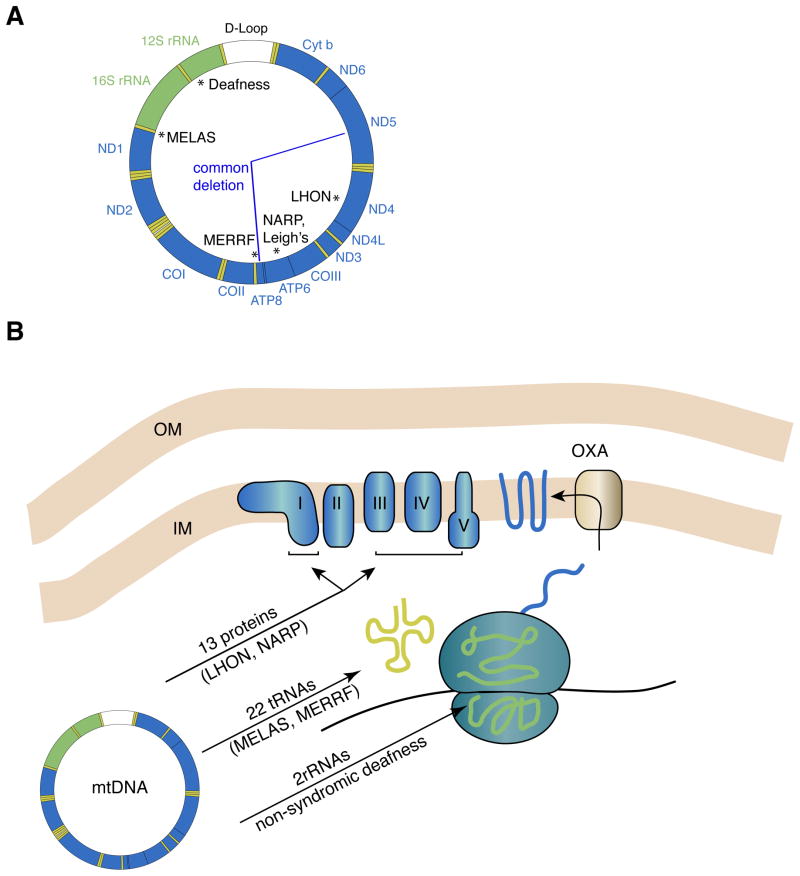
Figure 1. The human mtDNA genome and oxidative phosphorylation.
(A) Schematic of the circular mtDNA genome, showing the 13 protein coding genes (blue), the 2 rRNAs (green) and the 22 tRNAs (yellow). At the top is the non-coding D-loop (white), also known as the control region. This region is involved in mtDNA replication and transcriptional initiation. Classic examples of point mutations associated with prototypical mitochondrial encephalomyopathies are noted with asterisks. The “common deletion” removes 4977 bp of mtDNA and is one of many deletions that have been associated with sporadic KSS, PEO, and PS. (B) Oxidative phosphorylation and mtDNA gene products. The five enzyme complexes constituting the OXPHOS machinery reside in the mitochondrial inner membrane and consist of components encoded by both the nuclear and mitochondrial genomes. The 13 mtDNA proteins are transmembrane subunits of the enzyme complexes I, III, IV, and V. They are translated in the matrix of the mitochondrion and inserted into the inner membrane via the oxidase assembly (OXA) machinery. Mitochondrial ribosomes have polypeptides encoded by the nuclear genome. These polypeptides assemble into large and small ribosomal subunits that form complexes with rRNAs encoded by the mtDNA. The assembled ribosomes use mtDNA-encoded tRNAs to decode the messenger RNA. Examples of diseases caused by mutations in mtDNA-encoded proteins, tRNAs, and rRNAs are indicated.