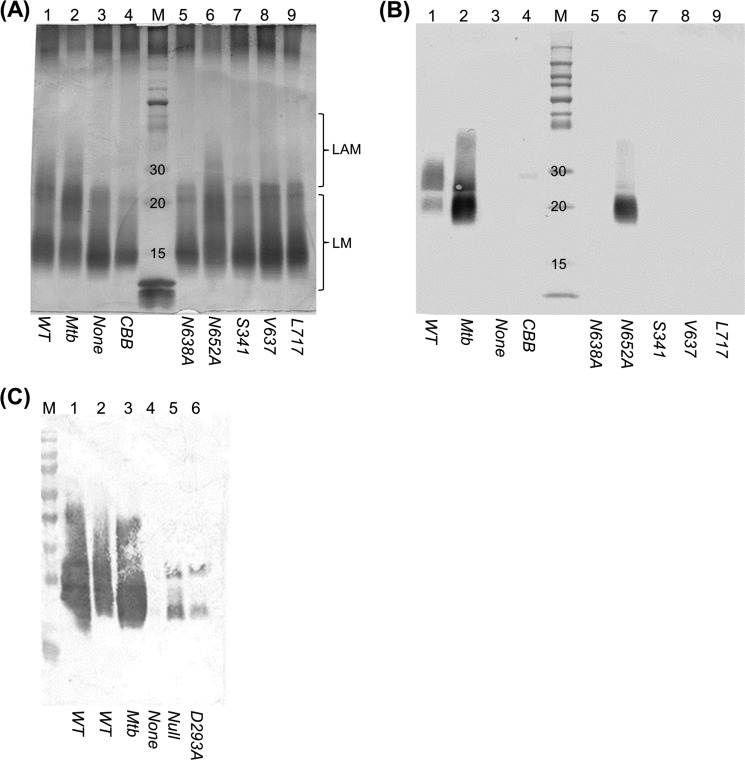
FIGURE 5.
Analysis of LAM from M. smegmatis recombinants. The M. smegmatis embCΔ deletion strain was complemented with the indicated embCMtb alleles. LAM was analyzed in wild-type and recombinant strains grown on agar. A, periodic acid-Schiff staining. B, Western blotting using antibody CS-35. Lane 1, wild-type M. smegmatis; lanes 2–9, M. smegmatis embCΔ complemented with wild-type embCMtb (lane 2), no allele (lane 3), embCBBMtb hybrid (lane 4), embCMtb N638A (lane 5), embCMtb N652A (lane 6), embCMtb Ser-341 truncation (lane 7), embCMtb Val-637 truncation (lane 8), and embCMtb Leu-717 truncation (lane 9); and lane M, markers. C, Western blotting using antibody CS-35. Lanes 1 and 2, wild-type M. smegmatis; lane 3, wild-type embCMtb; lane 4, empty; lane 5, no allele; and lane 6, embCMtb D293A. The expected range for migration of LAM and LM is shown for the PAS-stained gel. The Western blot detects only LAM.