Background: Hopanoids are present in bradyrhizobial lipid A preparations.
Results: Signals from hopanoid carboxyl shows strong correlation with the proton geminal to the hydroxy group of ester-linked long chain fatty acid.
Conclusion: Hopanoids are covalently linked to the lipid A of Bradyrhizobium.
Significance: The presence of such an unusual lipid A substituent may have a strong influence on the membrane properties of Bradyrhizobium.
Keywords: Glycoconjugate, Lipid A, Lipopolysaccharide (LPS), Mass Spectrometry (MS), Nuclear Magnetic Resonance (NMR), Bradyrhizobium, Hopanoid, Long Chain Fatty Acids
Abstract
The chemical structures of the unusual hopanoid-containing lipid A samples of the lipopolysaccharides (LPS) from three strains of Bradyrhizobium (slow-growing rhizobia) have been established. They differed considerably from other Gram-negative bacteria in regards to the backbone structure, the number of ester-linked long chain hydroxylated fatty acids, as well as the presence of a tertiary residue that consisted of at least one molecule of carboxyl-bacteriohopanediol or its 2-methyl derivative. The structural details of this type of lipid A were established using one- and two-dimensional NMR spectroscopy, chemical composition analyses, and mass spectrometry techniques (electrospray ionization Fourier-transform ion cyclotron resonance mass spectrometry and MALDI-TOF-MS). In these lipid A samples the glucosamine disaccharide characteristic for enterobacterial lipid A was replaced by a 2,3-diamino-2,3-dideoxy-d-glucopyranosyl-(GlcpN3N) disaccharide, deprived of phosphate residues, and substituted by an α-d-Manp-(1→6)-α-d-Manp disaccharide substituting C-4′ of the non-reducing (distal) GlcpN3N, and one residue of galacturonic acid (d-GalpA) α-(1→1)-linked to the reducing (proximal) amino sugar residue. Amide-linked 12:0(3-OH) and 14:0(3-OH) were identified. Some hydroxy groups of these fatty acids were further esterified by long (ω-1)-hydroxylated fatty acids comprising 26–34 carbon atoms. As confirmed by mass spectrometry techniques, these long chain fatty acids could form two or three acyloxyacyl residues. The triterpenoid derivatives were identified as 34-carboxyl-bacteriohopane-32,33-diol and 34-carboxyl-2β-methyl-bacteriohopane-32,33-diol and were covalently linked to the (ω-1)-hydroxy group of very long chain fatty acid in bradyrhizobial lipid A. Bradyrhizobium japonicum possessed lipid A species with two hopanoid residues.
Introduction
Lipopolysaccharide (LPS) is an integral component of most Gram-negative bacteria cell envelopes. LPS is usually composed of three domains: lipid A, a hydrophobic part that anchors the LPS molecule in the outer membrane and constitutes their outer leaflet, the core oligosaccharide, and very often the O-specific polysaccharide (O-chain). Such LPS is called smooth, found, for example, in Bradyrhizobium japonicum, Bradyrhizobium yuanmingense, and Bradyrhizobium sp. (Lupinus). LPS composed only of lipid A and the core oligosaccharide is called rough. The semi-rough form additionally containing one repeating unit of O-chain was found in Bradyrhizobium elkanii and Bradyrhizobium liaoningense strains (1). Bradyrhizobia are a slow-growing rhizobia forming a beneficial symbiosis with legumes. The endosymbiotic form of rhizobia, in which nitrogen fixation takes place, is called bacteroids. Rhizobial LPS plays an essential role in symbiosis progression. Together with membrane proteins and lipids favors optimal membrane architecture and determine its permeability, important for the morphology and functionality of bacteroids. Several reports demonstrated that the proper structure of rhizobial LPS is essential for root hair infection, nodule invasion, and adaptation to the endosymbiotic conditions (2–5). The LPS also protects microsymbiont cells against plant defense responses, i.e. hypersensitivity reaction and systemic acquired resistance, by suppressing such reactions during rhizobial infection (6–8).
LPS isolated from enterobacterial cells is often toxic, which is due to a certain lipid A structure. Toxic enterobacterial lipid A consists of a β-(1→6)-linked glucosaminyl disaccharide substituted by two phosphate groups at positions C-1 and C-4′. Six fatty acid residues, which form two acyloxyacyl moieties, are linked in distinct positions to the sugar backbone (9, 10). The activity of such lipid A results from binding to the TLR4-MD2 receptor complex on macrophages and endothelial cells (11). Activation of TLR4-MD2 initiates a signaling cascade, which results in biosynthesis of pro-inflammatory mediators by macrophages. In severe cases this may lead to septic shock (12). It was proven that, in humans, only such a lipid A structure is able to activate TLR4-MD2, determined by the set of fatty acids, their length, linkages, and spatial conformation (i.e. the presence of two acyloxyacyl residues) (10, 13). In lipid A samples, the structures of which differ from such enterobacterial ones, are usually less or not toxic in humans (1, 14, 15). The backbone of rhizobial lipid A can be composed either of a glucosaminyl-(d-GlcpN) or a 2,3-diamino-2,3-dideoxy-glucosaminyl-(d-GlcpN3N) disaccharide. In some Rhizobium species, the reducing residue of glucosamine-containing lipid A may be oxidized to 2-aminogluconate (16). The sugar backbone of lipid A can be decorated by phosphate, uronic acid, or mannose residues. The amino groups of d-GlcpN3N and d-GlcpN as well as C-3 and C-3′ of d-GlcpN are substituted by 3-hydroxy fatty acids. Hydroxyl groups of the primary fatty acids can be further substituted by nonpolar or very long chain (ω-1)-hydroxy VLCFAs,2 forming acyloxy-acyl moieties (17–21). The number of carbon atoms in VLCFA differs in each rhizobium strain. The 27-octacosanoic acid is present in LPS of all members of Rhizobiales, except of Azorhizobium caulinodans (22–24). Moreover, in bradyrhizobial LPSs several VLCFAs were identified, among them straight, and mono, and dimethyl branched-chain fatty acids built up of 26 to 34 carbon atoms (25). VLCFAs can span the entire outer membrane and play a crucial role in its stabilization.
The structure of lipid A isolated from B. elkanii USDA 76 has been described in detail recently (21). It differs considerably not only from other Gram-negative bacteria but also from other rhizobial lipid A samples. The sugar backbone is constituted by five sugar units identified as two diaminoglucoses and three mannoses. There are four amide-linked fatty acids (two 12:0(3-OH) and two 14:0(3-OH)) in this lipid A. Two of them are substituted by two VLCFA hydroxylated fatty acids, forming acyloxy-acyl residues. Additionally, one of them can be further acylated by 3-hydroxybutyric acid, linked to the (ω-1) hydroxy group. In this article we present the unique lipid A structure from different Bradyrhizobium strains, which form two or three acyloxyacyl moieties with VLCFAs and also contain one or two hopanoid residues linked covalently as tertiary hydrophobic residue(s) to the hydroxy group(s) of (ω-1)-hydroxy fatty acids.
Hopanoids are a class of pentacyclic triterpenoid lipids occurring in a wide range of Gram-negative and Gram-positive bacteria. Squalene and different species of hopanoid derivatives were discovered in Streptomyces spp., ethanol-tolerant bacterium Zymomonas mobilis, purple non-sulfur bacterium Rhodopseudomonas palustris, and Rhodomicrobium vanniellii, Methylococcus capsulatus, nitrogen-fixing Azotobacter and Beijerinckia, in symbiotic nitrogen-fixing Frankia and a wide group of Bradyrhizobium strains (26–30). Hopanoid lipids are thought to stabilize the phospholipid plasma membranes, sharing this function with eukaryotic sterols (31). In nitrogen-fixing bacteria this lipid component may have additional functions, besides membrane reinforcement. It has been proven that in Frankia, hopanoids can be involved in oxygen protection of the nitrogenase complex by forming of a diffusion barrier (27). In the case of Rh. palustris the bacteriohopane polyols determine membrane integrity and play a role in pH homeostasis (30). Very recently, the first hopanoid-containing lipid A, obtained from LPS of the photosynthetic Bradyrhizobium strain BTAi1, was structurally and functionally characterized (32).
EXPERIMENTAL PROCEDURES
Bacterial Strains and Culture Condition
Bacteria (B. japonicum USDA 110, B. yuanmingense CCBAU 10071, and Bradyrhizobium sp. (Lupinus) USDA 3045) were grown at 28 °C in 79CA medium according to Vincent (33), for 14 days, with aeration by vigorous shaking.
Isolation and Purification of LPS and Lipid A Samples
The cell pellets obtained by centrifugation were washed twice with saline, once with distilled water, and then delipidation was performed according to Que and co-workers (19). The delipidated and dried cell pellets were suspended in 50 mm sodium phosphate buffer (pH 7.0), supplemented with 5 mm EDTA, and digested with lysozyme (6 mg g−1 dry mass, 4 °C, 16 h). The nucleic acids were degraded by treatment with DNase and RNase (0.3 mg g−1 dry mass, 37 °C, 30 min). Cell proteins were digested by incubation with proteinase K (0.3 mg g−1 dry mass, room temperature, for 18 h, followed by incubation for 10 min at 60 °C) (34). The LPS preparations were obtained from hot 45% phenol/water extractions according to Westphal and Jann (35), with further modifications (36). The phenol and water phases, which contained LPS, were dialyzed extensively against tap and distilled water. Pure LPS preparations were obtained by ultracentrifugation (105,000 × g, 4 °C, 4 h). The LPS was obtained from water phase after phenol/water extraction, 820 mg (5.8%) in the case of B. japonicum, 148 mg (1.4%) in the case of B. yaunmingense, and 344 mg (5.7%) in the case of Bradyrhizobium sp. (Lupinus).
Lipid A was liberated from LPS by mild acid hydrolysis (1–2% aqueous acetic acid, 100 °C, 2–3 h). The free lipid A was purified by a two-phase Bligh-Dyer system according to Que et al. (19). Briefly, adequate amounts of chloroform and methanol were added to the hydrolysate to obtain a chloroform/methanol/hydrolysate, 2:2:1.8 (v/v/v), mixture. The mixture was vigorously shaken and then centrifuged. The chloroform phase, containing lipid A, was collected and washed twice with the water phase from the freshly prepared two-phase Bligh-Dyer mixture (chloroform/methanol/water, 2:2:1.8 (v/v/v)). The pure lipid A preparations (B. japonicum, 36 mg; B. yuanmingense, 18 mg; Bradyrhizobium sp. (Lupinus), 12 mg) were stored at −20 °C in CHCl3/MeOH (3:1, v/v). O-Deacylation of lipid A samples was performed by incubation (1–2 mg) in chloroform, methanol, 0.6 m aqueous NaOH, 2:3:1 (v/v/v), for 1.5 h at room temperature, according to Que-Gewirth and co-workers (37).
Fatty Acids, Hopanoid Lipids, and Sugars Analysis
For total fatty acid and hopanoid lipids determination, lipid A preparations were subjected to hydrolysis in 4 m HCl (100 °C, 4 h). Liberated fatty acids and hopanoids were extracted with chloroform and converted to their methyl esters with diazomethane. After evaporation to dryness, hydroxyl groups of fatty acids and hopanoid lipids were derivatized with BSTFA (16 h at room temperature). Neutral and amino sugar analyses were performed according to standard protocols described elsewhere (21).
GC-MS analyses of fatty acids and sugars were performed on a Hewlett Packard gas chromatograph 5890 series II and Agilent Technologies GC System 7890A connected to a mass selective detector EI/CI MSD 5975C, equipped with a HP-5MS column (30 m × 0.25 mm) with helium as a carrier gas (flow rate: 0.7 ml min−1). The temperature program was as follows: 150 °C for 3 min, then raised to 250 °C at 3 °C min−1, then to 320 °C, 25 °C min−1. The final temperature was kept for 10 min for sugar analysis and 20 min for fatty acid analysis.
Mass Spectrometry
Lipid A samples obtained from B. japonicum were analyzed on a high resolution hybrid Fourier transform ion cyclotron resonance mass spectrometry (FT-ICR) instrument (Apex Qe Bruker Daltonics, Billerica, MA) with electrospray ionization (ESI), equipped with a 7 tesla actively shielded magnet. Samples for analysis were prepared as described earlier (21) and measured in the negative ion mode. Mass spectra were charge deconvoluted and mass numbers given refer to the monoisotopic peaks.
Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF-MS) was performed with a Bruker-Daltonics Reflex III instrument (Bruker Daltonics, Bremen, Germany) at an acceleration voltage of 20 kV and delayed ion extraction. Lipid A preparations were dispersed in methanol/water (1:1, v/v) at a concentration of 1 μg/μl with addition of 20 mm EDTA. The matrix solution was prepared from 2,5-dihydroxybenzoic acid in 1% trifluoroacetic acid and the spectra were recorded in positive or negative ion modes.
NMR Spectroscopy
For NMR analysis a sample containing 18 mg of native lipid A from B. japonicum dissolved in 0.6 ml of CDCl3/CD3OD (2:1, v/v) with 5 μl of D2O, was used. One- and two-dimensional NMR spectra were recorded at 700 MHz on an AVANCE III spectrometer with Cryoprobe (Bruker) using Bruker software. Spectra were recorded at 27 °C. The following two-dimensional NMR experiments were performed: COSY, DQF-COSY, TOCSY, ROESY, HSQCnd, HSQC-DEPT, and HMBC. The 1H and 13C resonances were measured relative to TMS (δH 0.0/δC 0.0).
RESULTS
Isolation and Chemical Analysis of Lipid A of Bradyrhizobium
LPS of Bradyrhizobium strains were isolated using hot phenol/water extraction. The material was found mainly in the water phase (for yields see ”Experimental Procedures“). Lipid A was liberated from LPS by mild acid hydrolysis and subsequently subjected to sugar and fatty acid analyses. d-GlcpN3N was the only amino sugar component. A high amount of mannose (d-Manp) was also present in all lipid A preparations, whereas only small amounts of galacturonic acid (d-GalpA) were detected. Fatty acid analysis revealed the presence of two amide-linked 3-hydroxy fatty acids, i.e. 12:0(3-OH) and 14:0(3-OH). Ester-linked VLCFAs with chain lengths of 26–34 carbon atoms were identified as well (Table 1). The most abundant VLCFAs of strain B. japonicum were 26:0(25-OH), 32:0(31-OH), and 33:0(32-OH). Lipid A from B. yuanmingense and Bradyrhizobium sp. (Lupinus) contained mainly 31:0(30-OH).
TABLE 1.
Fatty acid, hopanoids, and sugar components of lipid A isolated from LPS of Bradyrhizobium strains
The symbols represent: +, present; ++, the main fatty acid/sugar component; −, lack of component; tr, traces.
| Component | B. japonicum USDA 110 | B. yuanmingense CCBAU 10071 | Bradyrhizobium sp. (Lupinus) USDA 3045 |
|---|---|---|---|
| Fatty acids | |||
| 12:0(3-OH) | ++ | ++ | ++ |
| 14:0(3-OH) | ++ | ++ | ++ |
| 26:0(25-OH) | ++ | + | + |
| 27:0(26-OH)a | − | tr. | tr. |
| 28:0(27-OH) | + | + | + |
| 29:0(28-OH)a | + | − | + |
| 30:0(29-OH) | + | + | + |
| 31:0(30-OH)a | ++ | ++ | ++ |
| 32:0(31-OH)b | ++ | − | + |
| 33:0(32-OH)a | ++ | + | ++ |
| 34:0(33-OH) | − | + | − |
| Hopanoidsc | |||
| I, R = CH3 | + | + | + |
| II, R = H | ++ | ++ | ++ |
| Sugars | |||
| d-Manp | ++ | ++ | ++ |
| d-GalpA | + | + | + |
| d-GlcpN3N | + | + | + |
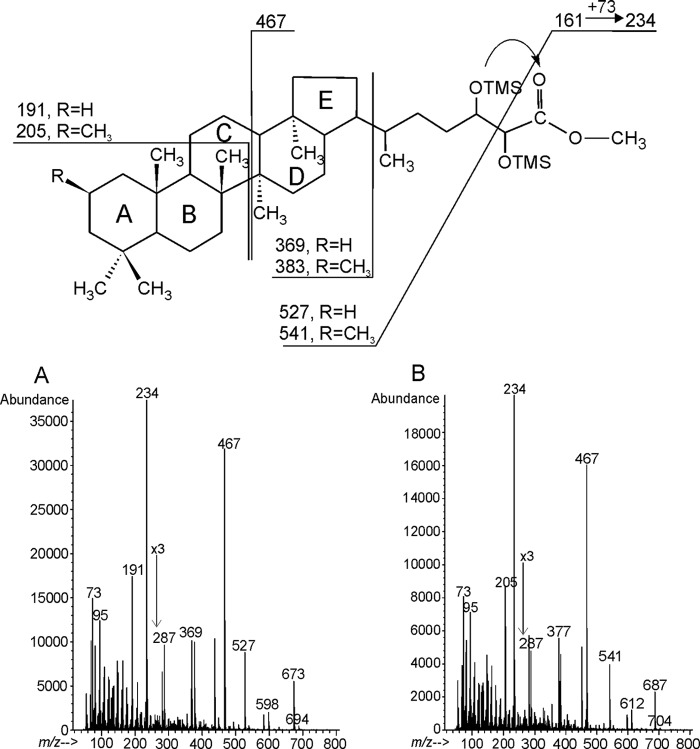
In all lipid A preparations a high amount of pentacyclic triterpenoids (hopanoid lipids) was present. Based on GC-MS data of trimethylsilyloxy derivatives of methyl esters obtained from the B. japonicum lipid A preparation two constituents were identified (Fig. 1, A and B). The characteristic fragmentations defined them as 34-carboxyl-bacteriohopane-32,33-diol, the major component of bradyrhizobial lipid A, and 34-carboxyl-2-methyl-bacteriohopane-32,33-diol as the less abundant component. The dominating signal at m/z 234 visible on both mass spectra derived from the cleavage between C-33 and C-32 and corresponded to the fragment bearing a carboxyl group. This ion resulted from a rearrangement followed by transferring the TMS group from position C-32 to the carboxyl group. The mass spectrum of the TMS derivative of 34-carboxyl-bacteriohopane-32,33-diol methyl ester (tR = 50.645 min, Fig. 1A) showed a pseudomolecular ion at m/z 673, [M-15]+, and a prominent ion at m/z 191, which were derived from fragmentation of the third (C) ring of the molecule. The mass spectrum of the TMS derivative of 34-carboxyl-2-methyl-bacteriohopane-32,33-diol methyl ester (tR = 50.084 min, Fig. 1B) possessed [M-15]+ at m/z 687 and a C-ring fragmentation that yielded an ion at m/z 205 (38). The presence of the last ion suggested that an additional methyl group could be localized in rings A or B. It is known that 3-methylhopanoids elute later than the non-methylated species, however, in the case of 2-methylhopanoids the retention time is very close to the corresponding nonmethylated ones (39). In B. japonicum lipid A, the TMS derivative of the methylated hopanoid eluted a bit earlier than the non-methylated form, suggesting the presence of a methyl group at C-2 of ring A (Fig. 1).
FIGURE 1.
Mass spectra (GC-MS) of hopanoid components of B. japonicum lipid A (acid hydrolyzed, carboxymethylated, and trimethylsilylated preparation). A, TMS derivative of 34-carboxyl-bacteriohopane-32,33-diol methyl ester; B, TMS derivative of 34-carboxyl-2β-methyl-bacteriohopane-32,33-diol methyl ester.
Mass Spectrometry of Lipid A Preparations
Lipid A preparations were investigated either by ESI FT-ICR-MS or MALDI-TOF-MS.
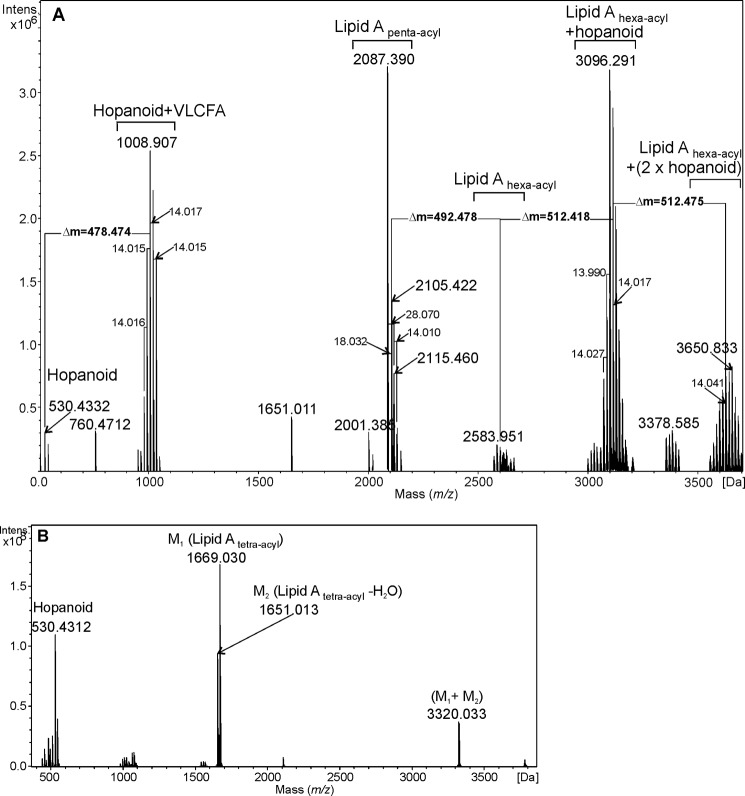
The charge-deconvoluted ESI FT-ICR mass spectrum of the native lipid A of B. japonicum showed lipid A molecules comprising a different acylation pattern, which can be recognized by the mass difference of 14 and 28 Da between neighboring signals (Fig. 2A and Table 2). Monoisotopic masses 2087.390, 2105.422, and 2115.460 Da were assigned to lipid A species containing two Manp, two GlcpN3N, one GalpA, two 12:0(3-OH), two 14:0(3-OH), and one ester-linked fatty acid, forming penta-acyl lipid A. The mass difference of 18 Da originated from a dehydration process, occurring during cleavage of VLCFA. The cluster of low-intensity signals in the 2570–2680 Da region was derived from hexa-acylated lipid A molecules containing two secondary VLCFA substituents. The intensive peaks at 3096.291 and 3110.318 Da could be assigned to the hexa-acylated lipid A that contained two ester-linked VLCFA, like 29:0(28-OH) and 32:0(31-OH) or 29:0(28-OH) and 33:0(32-OH). It was postulated that one of those VLCFAs was linked to the hopanoid residue (Δm = 512.418 Da) via its hydroxyl group. Such lipid A molecules possess a calculated monoisotopic mass of 3096.343 and 3110.358 Da. Mass differences of 14 Da were due to different lengths of VLCFAs as well as the presence of two hopanoid species. Signals derived from molecules with the highest mass (around 3600 Da) originated from hexa-acyl lipid A containing two hopanoid substituents as tertiary residues, moreover, one of these hopanoid moieties could bear a 2β-methyl group (see Fig. 1).
FIGURE 2.
Charge-deconvoluted ESI FT-ICR mass spectrum of the native (A) and O-deacylated (B) lipid A isolated from B. japonicum.
TABLE 2.
Annotation of mass spectrometric information (main signals) obtained for the O-deacylated and native lipid A samples from B. japonicum (see Fig. 2, A and B)
| Form of lipid A | Measured mass | Type of molecule | Composition | Calculated monoisotopic mass |
|---|---|---|---|---|
| Da | Da | |||
| Lipid A O-deacylated | 530.4312 | Hopanoid | 530.4335 | |
| 1669.030 | Lipid Atetra-acyl | 2 × GlcpN3N | 1669.0174 | |
| 2 × Manp | ||||
| 1 × GalpA | ||||
| 2 × 14:0 (3-OH), 2 × 12:0 (3-OH) | ||||
| -H2O | ||||
| Lipid A native | 760.4712 | [Y]-ion type | 1 × GlcpN3N | 760.4726 |
| 1 × GalpA | ||||
| 1 × 14:0 (3-OH), 1 × 12:0 (3-OH) | ||||
| 1008.907 | Hopanoid | Hopanoid 530.4335 u | 1008.9084 | |
| + VLCFA | 1 × 32:0 (31-OH) | |||
| 1651.011 | Lipid Atetra-acyl | 2 × GlcpN3N | 1651.0068 | |
| -H2O | 2 × Manp | |||
| 1 × GalpA | ||||
| 2 × 14:0 (3-OH), 2 × 12:0 (3-OH) | ||||
| −2 × H2O | ||||
| 2087.390 | Lipid Apenta-acyl | 2 × GlcpN3N | 2087.4353 | |
| -H2O | 2 × Manp | |||
| 1 × GalpA | ||||
| 2 × 14:0 (3-OH), 2 × 12:0 (3-OH) | ||||
| 1 × 29:0 (28-OH) | ||||
| − 2 × H2O | ||||
| 2583.951 | Lipid Ahexa-acyl | 2 × GlcpN3N | 2583.920 | |
| 2 × Manp | ||||
| 1 × GalpA | ||||
| 2 × 14:0 (3-OH), 2 × 12:0 (3-OH) | ||||
| 1 × 29:0 (28-OH), 1 × 32:0 (31-OH) | ||||
| -H2O | ||||
| 3096.291 | Lipid Ahexa-acyl | 2 × GlcpN3N | 3096.3428 | |
| + hopanoid | 2 × Manp | |||
| 1 × GalpA | ||||
| 2 × 14:0 (3-OH), 2 × 12:0 (3-OH) | ||||
| 1 × 29:0 (28-OH), 1 × 32:0 (31-OH) | ||||
| 1 × hopanoid 530.4335 | ||||
| -H2O | ||||
| 3650.833 | Lipid Ahexa-acyl | 2 × GlcpN3N | 3650.8126 | |
| + (2 × hopanoid) | 2 × Manp | |||
| 1 × GalpA | ||||
| 2 × 14:0 (3-OH), 2 × 12:0 (3-OH) | ||||
| 1 × 31:0 (30-OH), 1 × 32:0 (31-OH) | ||||
| 1 × hopanoid 530.4335 | ||||
| 1 × hopanoid 544.4491 | ||||
| -H2O |
Mass peaks around 1000 Da originated either from the hopanoid-VLCFA moiety that was cleaved from the native lipid A during mild acid hydrolysis or could be the result of fragmentation during ionization. The mentioned dehydrated form of penta-acylated lipid A (2087.390 Da) likely also resulted from this process. The mass differences between neighboring peaks in this cluster equal 14 Da, originating from both, the different lengths of linked VLCFA and the methylated form of the hopanoid.
The mass spectrum of O-deacylated lipid A of B. japonicum USDA 110 contained three sets of signals (Fig. 2B). The peaks at 530.4312 Da were derived from a hopanoid residue, which was cleaved during O-deacylation and was not removed by extraction. The mass peaks at 1651.013 and 1669.030 Da were derived from the tetra-acylated lipid A. The second signal was consistent with a lipid A species composed of two GlcpN3N, two Manp, one GalpA, and four amide-linked fatty acid residues (two 12:0(3-OH) and two 14:0(3-OH)). One 3-OH fatty acid was deprived of H2O resulting in an α-unsaturated derivative (see the text above). The signal at 1651.013 Da corresponded to a lipid A built from the same components, which unspecifically lost another water molecule (Δm = 18 Da). The group of peaks at 3320.033 Da was consistent with the ion-cluster of both forms of tetra-acyl lipid A.
Fig. 3, A and B, shows MALDI-TOF mass spectra (positive ion mode) obtained on the native and O-deacylated lipid A preparations isolated from B. yuanmingense. Three sets of ions are visible on the spectrum of native lipid A. The main signals at m/z 2669.1, 3133.4, and 3660.6 were consistent with calculated masses of hexa-acylated lipid A, hepta-acylated lipid A, and hepta-acylated-hopanoid containing lipid A, respectively. The mass spectrum of the O-deacylated sample (Fig. 3A) showed a main signal at m/z 1692.3, originating from sodiated tetra-acyl lipid A, composed of two GlcpN3N units, two Manp, one GalpA, and four amide-linked fatty acids (two 12:0(3-OH) and two 14:0(3-OH) −H2O). The calculated monoisotopic mass for the [M + Na]+ ion was determined to m/z 1692.0. The signals identified as hexa-acyl lipid A derived from molecules containing two ester-linked VLCFAs (such as 33:0(32-OH) and 34:0(33-OH), with a calculated [M-H2O + H]+ m/z of 2669.02, Fig. 3B). The ion cluster at m/z 3100–3200 was identified as hepta-acyl lipid A containing a third acyl residue (such as 31:0(30-OH), theoretical m/z of [M-H2O + H]+ = 3133.48). The lipid A molecules with mass around 3660 Da additionally contained one hopanoid residue, ester-linked as in lipid A from B. japonicum to the (ω-1)-OH-group of one of the VLCFA residues.
FIGURE 3.
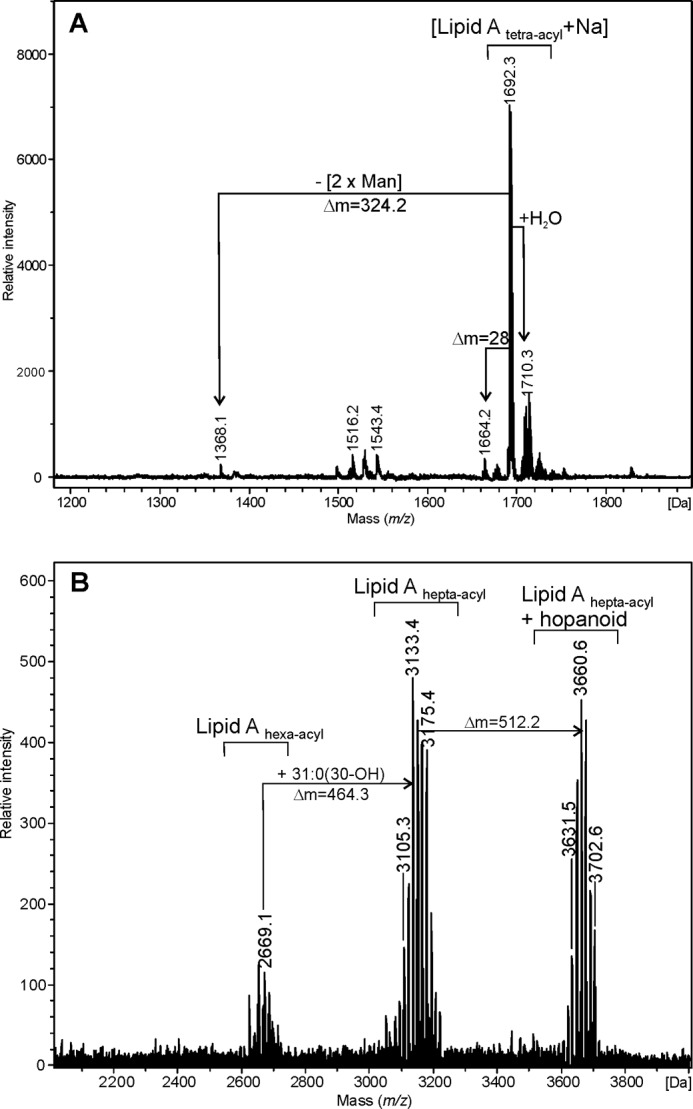
MALDI-TOF mass spectrum (positive-ion mode) of the O-deacylated (A) and native (B) lipid A from B. yuanmingense.
Fig. 4 shows the MALDI-TOF mass spectrum (negative ion mode) of the lipid A from Bradyrhizobium sp. (Lupinus). Two sets of signals are visible, one around m/z 2583.0 and the other at m/z 3095.2, which were assigned to hexa-acyl lipid A carrying two VLCFAs (e.g. 30:0(29-OH) and 31:0(30-OH), theoretical m/z for [M-H2O − H]− = 2582.91) and a hexa-acylated lipid A moiety additionally bearing one hopanoid residue (calculated m/z [M-H2O − H]− = 3095.33).
FIGURE 4.
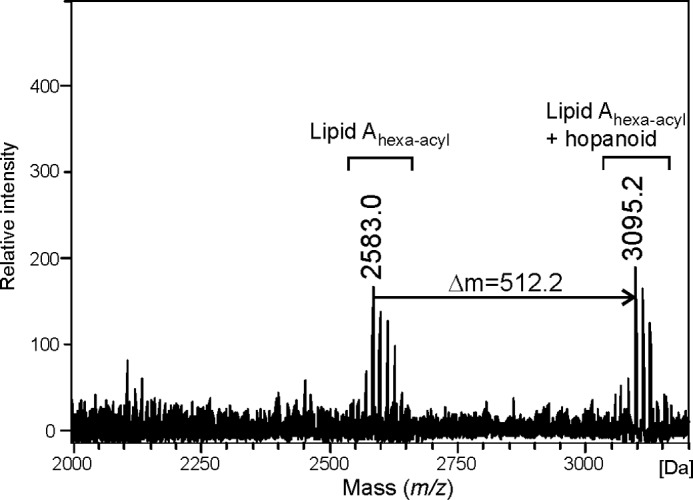
MALDI-TOF mass spectrum (negative-ion mode) of the native lipid A sample from Bradyrhizobium sp. (Lupinus).
NMR Spectroscopy of B. japonicum Lipid A
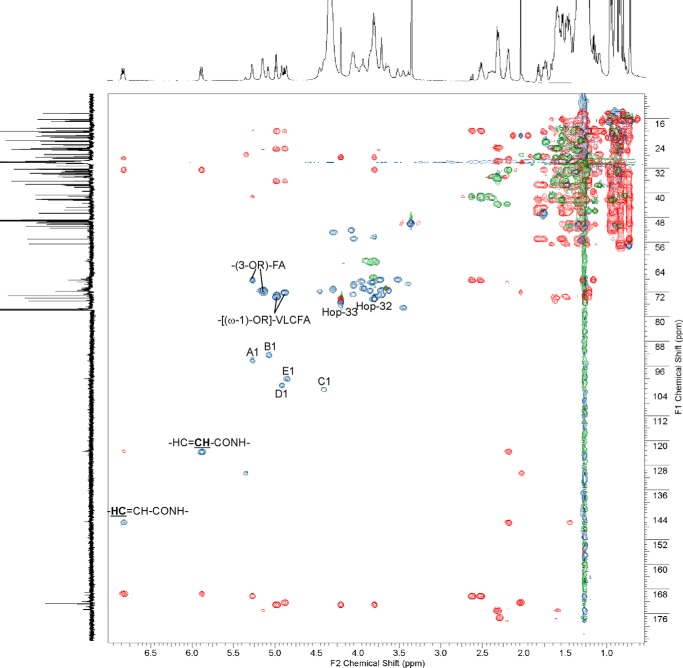
The native lipid A preparation isolated from B. japonicum was dissolved in CDCl3/CD3OD (2:1, v/v) with traces of D2O and fully characterized by one- and two-dimensional NMR spectroscopy. Lipid A was not substituted by phosphate residues, as confirmed by 31P NMR spectroscopy. The HSQC-DEPT spectrum of the lipid A (Fig. 5, blue and green) contained five signals of anomeric carbons (δ 92.25–103.14), four signals of nitrogen-bearing carbons (δ 52.02–54.49), those of sugar ring carbon atoms (δ 61.47–76.76) and signals for CH-OR as well as for CH-OH groups from fatty acids and hopanoid lipids (δ 68.1–73.2).
FIGURE 5.
HSQC-DEPT (blue and green) and HMBC (red) spectra of native lipid A from B. japonicum. Lipid A sample was dissolved in CDCl3/CD3OD (2:1, v/v) with traces of D2O. The signals were marked as follows: A, α-d-GalpA; B, α-d-GlcpN3N; C, β-d-GlcpN3N; D, α-d-Manp; E, α-d-Manp; Hop-32 and Hop-33, CH-OH groups at positions of 32 and 33, respectively, of hopanoid lipid side chain (see Fig. 4).
Based on 1H/1H COSY, TOCSY, and 1H/13C HMBC experiments five spin systems characterizing sugar pyranoses were identified. Two of them (E and D) were derived from α-d-Manp, C represented β-d-GlcpN3N, B represents α-d-GlcpN3N, and A was α-d-GalpA. All 1H and 13C chemical shifts for lipid A sugar backbone components were assigned and are listed in Table 3. The anomeric configuration of monosaccharides was confirmed by measuring 1J(C1,H1) coupling constants. Relatively large values of coupling constants (above 170 Hz) for anomeric signals were found for residues A, B, D, and E, thus identifying their α-configuration. A smaller value of 1J(C1,H1) (∼164 Hz) was found for residue C, determining its β-configuration. The following connectivities between anomeric and linkage protons were identified on ROESY spectrum: A1/B1 (δ 5.270/5.078), C1/B6a,b (δ 54.407/3.802 and 4.407/3.662), D1/C4 (δ 4.910/3.653), and E1/D6 (δ 4.854/3.816). Taken together, the sugar backbone of B. japonicum lipid A possessed the structure: α-d-Manp-(1→6)-α-d-Manp-(1→4)-β-d-GlcpN3N-(1→6)-α-d-GlcpN3N-(1→1)-α-d-GalpA.
TABLE 3.
1H and 13C NMR signals (δ; ppm) from B. japonicum lipid A backbone (in CDCl3/CD3OD (2:1) with traces of D2O)
13C chemical shifts of atoms involved in glycosidic linkages are bold.
| Spin system | 1J(C-1,H-1) | H-1/C-1 | H-2/C-2 | H-3/C-3 | H-4/C-4 | H-5/C-5 | H-6/C-6 | H-6′ |
|---|---|---|---|---|---|---|---|---|
| Hz | ||||||||
| A (α-d-GalpA) | 177.6 | 5.270 | 3.966 | 4.050 | 4.291 | 4.459 | ||
| 94.00 | 68.30 | 69.63 | 70.96 | 71.54 | 169.3 | |||
| B (α-d-GlcpN3N) | 176.4 | 5.078 | 4.077 | 4.290 | 3.395 | 4.055 | 3.802 | 3.662 |
| 92.25 | 52.02 | 52.54 | 69.22 | 72.56 | 69.96 | |||
| C (β-d-GlcpN3N) | 163.9 | 4.407 | 3.805 | 4.046 | 3.653 | 3.447 | 3.788 | 3.900 |
| 103.14 | 54.09 | 54.49 | 75.86 | 76.76 | 61.47 | |||
| D (α-d-Manp) | 172.0 | 4.910 | 3.707 | 3.628 | 3.515 | 3.712 | 3.816 | 3.816 |
| 101.96 | 71.17 | 71.27 | 67.66 | 72.83 | 67.17 | |||
| E (α-d-Manp) | 172.2 | 4.854 | 3.934 | 3.858 | 3.715 | 3.779 | 3.861 | 3.786 |
| 99.94 | 70.68 | 71.54 | 67.62 | 73.19 | 61.98 |
The fine structure of both hopanoid components of bradyrhizobial lipid A was identified. Carbon signals characteristic for the main hopanoid residue in lipid A are listed in Table 4. In the HSQC-DEPT spectrum (Fig. 5, blue and green), the hopanoids' ring, fatty acid bulk, and terminal signals grouped in the crowded region δH 0.7–2.8 and δC 16–57 ppm. Signals for CH-OH groups from positions 32 and 33 of the hopanoid side chains were located in the glycosidic region, at δ 3.800/73.99 and δ 4.200/74.94, respectively. The signal of the carboxyl group of the hopanoid was assigned at δC 172.73, and revealed a distinct correlation with the (ω-1) proton of VLCFA (CH-[(ω-1)-OR]-fragment, δH 4.980). Thus, the hopanoid moiety was a constitutive component of B. japonicum lipid A.
TABLE 4.
13C NMR chemical shifts of the hopanoid residue (34-carboxyl-bacteriohopane-32,33-diol) of B. japonicum lipid A
| Carbon atom | δ | |
|---|---|---|
| ppm | ||
| 1 | -CH2 | 40.66 |
| 2 | -CH2 | 19.02 |
| 3 | -CH2 | 42.40 |
| 4 | -C | 33.50 |
| 5 | -CH | 56.62 |
| 6 | -CH2 | 19.01 |
| 7 | -CH2 | 33.42 |
| 8 | -C | 42.00 |
| 9 | -CH | 50.00 |
| 10 | -C | 37.78 |
| 11 | -CH2 | 20.51 |
| 12 | -CH2 | 24.28 |
| 13 | -CH | 50.10 |
| 14 | -C | 41.90 |
| 15 | -CH2 | 33.98 |
| 16 | -CH2 | 23.27 |
| 17 | -CH | 55.06 |
| 18 | -C | 44.73 |
| 19 | -CH2 | 41.99 |
| 20 | -CH2 | 28.06 |
| 21 | -CH | 46.45 |
| 22 | -CH | 37.22 |
| 23 | -CH3 | 33.61 |
| 24 | -CH3 | 21.73 |
| 25 | -CH3 | 16.16 |
| 26 | -CH3 | 16.86 |
| 27 | -CH3 | 16.70 |
| 28 | -CH3 | 16.28 |
| 29 | -CH3 | 20.58 |
| 30 | -CH2 | 32.51 |
| 31 | -CH2 | 28.52 |
| 32 | -CH-OH | 73.99 |
| 33 | -CH-OH | 74.94 |
| 34 | -COOR*a | 172.73 |
a R*, lipid A (-(ω-1)-OH-group).
Position of the methyl group in 34-carboxyl-2-methyl-bacteriohopane-32,33-diol was confirmed based on HMBC, TOCSY, and ROESY correlations. A few changes were noticed in chemical shifts of carbons of rings A and B, compared with the non-methylated component. The carbon chemical shifts were as follows: δ 50.22 (C-1), 25.04 (C-2, methine group), 23.15 (2β-CH3), 45.45 (C-3), 46.51 (C-4), 50.00 (C-5), 32.87 (C-6), 19.95 (C-7), 41.92 (C-8), 31.23 (C-23), 26.28 (C-24), and 22.30 (C-25). As the carbon atom from the methyl group at C-2 only correlated with the proton at C-2 in the HMBC spectrum, this group should be in the β position (29). Also, the through-space connectivity of proton at C-2 with the protons from the methyl group confirmed its position as 2β. Moreover, protons from the methyl group showed correlation with protons of methyl groups at position C-24 and C-25 in the ROESY spectrum, but there was no correlation with protons at position C-23 (data not shown). Thus, evidence for β-configuration of this substituent was provided.
All chemical shifts of the α, β, and γ carbon and proton signals of the 3-hydroxy fatty acids (both, 3-O-acylated and those with free OH group) as well as for signals derived from ω, ω-1, ω-2, and ω-3 protons and carbons of substituted and unsubstituted VLCFA, are summarized in Table 5. Chemical shift data were similar to those reported for B. elkanii lipid A (21). The 1H/13C signals of the β-CH group of the unsubstituted 3-hydroxy fatty acid were identified at δ 3.82/68.88, respectively. Two signals derived from β-CH of 3-O-substituted fatty acids were found at δ 5.269/68.10 and 5.145/71.59. The proton/carbon chemical shifts at δ 4.98/73.21 and 4.88/72.07 were derived from the ω-1 methine groups of the VLCFAs, which are connected to the 3-OH group of the amide-linked fatty acids and bore hopanoid residue(s).
TABLE 5.
1H and 13C NMR chemical shifts of fatty acids from B. japonicum lipid A
| No. | Fatty acids signals | 1H | 13C |
|---|---|---|---|
| ppm | |||
| 1. | Olefinic protons/carbons | ||
| -CONH- | 169.28 | ||
| -HC = CH-CONH- | 6.823 | 146.19 | |
| -HC = CH-CONH- | 5.882 | 123.39 | |
| -CH2-CH2-HC = CH-CONH- | 2.184 | 32.60 | |
| -CH2-CH2-HC = CH-CONH- | 1.445 | 28.69 | |
| 2. | Olefinic protons/carbons (separated one double bound) | ||
| -CH2-HC = CH- | 5.348 | 130.26 | |
| -CH2-HC = CH- | 2.020 | 27.41 | |
| 3. | Ist –(3-OR′)-FAa | ||
| α1/α2 | 2.628/2.532 | 41.04 | |
| β | 5.269 | 68.10 | |
| γ | NDb | ND | |
| CONH-Sug | 169.96 | ||
| R-COO- | 164.57 | ||
| ω | 1.214 | 19.91 | |
| 4. | IInd – (3-OR″-FAa | ||
| α1/α2 | 2.413/2.525 | 41.03 | |
| β | 5.145 | 71.59 | |
| γ | 1.571 | 34.69 | |
| -CONH-Sug | 172.23 | ||
| R-COO- | 174.66 | ||
| 5. | Ist – [(ω-1)-OR*]c VLCFA | ||
| ω | 1.257 | 20.03 | |
| ω-1 | 4.980 | 73.21 | |
| ω-2 | 1.504; 1.623 | 36.14 | |
| ω-3 | 1.338 | 25.85 | |
| ω-4 and next CH2 groups | 1.450 | 28.91 | |
| R*(-COO-) from hopanoid | 172.82 | ||
| 6. | IInd – [(ω-1)-OR**]c VLCFA | ||
| ω | 1.213 | 20.03 | |
| ω-1 | 4.881 | 72.070 | |
| ω-2 | 1.487; 1.588 | 36.340 | |
| ω-3 | 1.308 | 25.67 | |
| R**(-COO-) from 2nd hopanoid | 172.00 | ||
| 7. | (3-OH) FA with unsubstituted OH group | ||
| α1/α2 | 2.310/2.210 | 43.81 | |
| β | 3.825 | 68.88 | |
| γ | ND | ND | |
| 8. | [(ω-1)-OH] VLCFA with unsubstituted OH group | ||
| ω | ND | ND | |
| ω-1 | 3.746 | 68.45 | |
| ω-2 | 1.386; 1.473 | 39.33 | |
| ω-3 | 1.267 | 26.10 | |
| 9. | [(ω-1)-OH] VLCFA with unsubstituted OH group | ||
| ω-1 | 3.575 | 67.61 | |
| ω-2 | 1.323; 1.435 | 33.19 | |
| ω-3 | 1.267 | 26.10 | |
a R′ and R″, VLCFA.
b ND, not determined.
c R* and R**, hopanoid.
Olefinic chemical shifts were identified at δ 6.82/146.19 and 5.88/123.39. HMBC (Fig. 5, red) and ROESY spectra indicated that the double bond was located at the α-C-atom of the amide-linked fatty acid. The formation of such α-unsaturated fatty acids was probably a result of the cleavage of VLCFAs ester-bound to O-3 of an amide-linked 3-OH fatty acid during prolonged mild acid liberation of lipid A from LPS. Thus, these α-unsaturated amide-linked fatty acids were artifacts originating from chemical treatment of the LPS. In addition, a signal characteristic for olefinic proton/carbon belonging to another double bond was also identified (δ 5.34/130.26). This signal is characteristic for unsaturated fatty acid possessing a double bond located in the so-called envelope of the acyl chain. Based on MS data it was concluded that presumably this fatty acid was not a component of lipid A and originated from traces of (phospho)lipids co-extracted with LPS. In summary, all data (GC-MS, NMR, and FT-ICR MS) obtained for B. japonicum lipid A identify its structure as depicted in Fig. 6.
FIGURE 6.
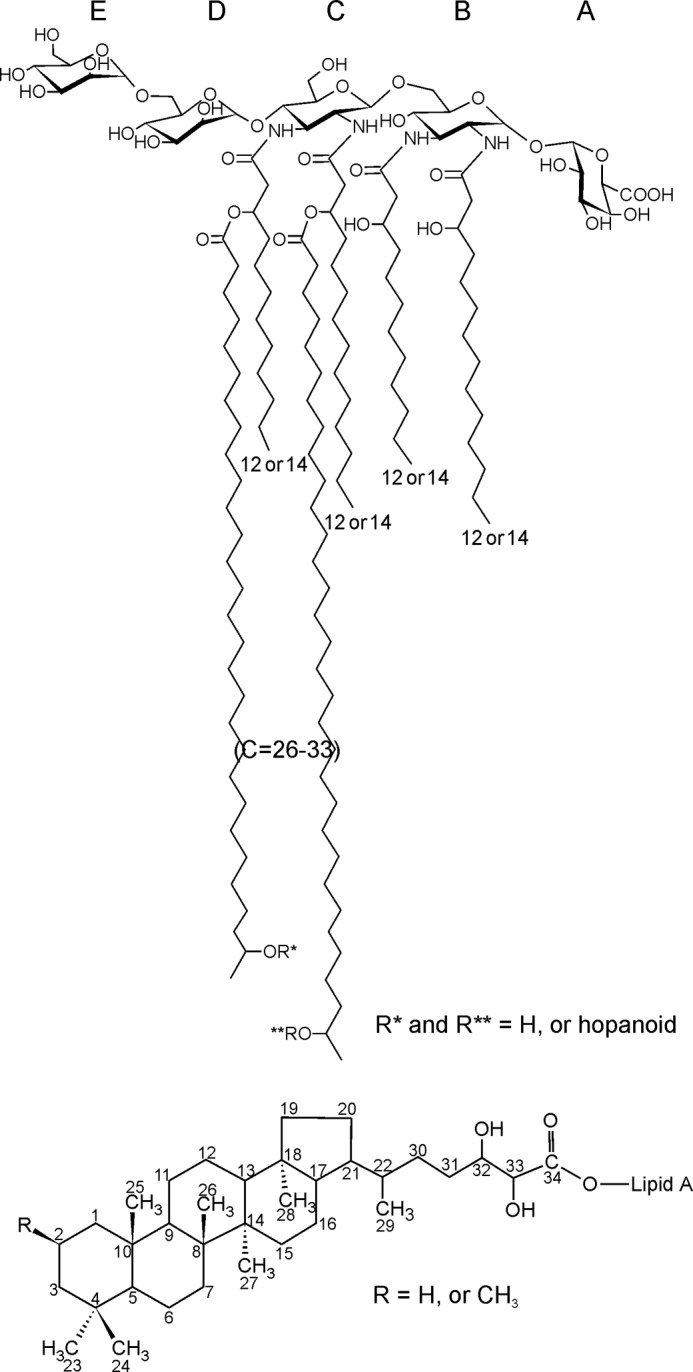
Proposed structure of B. japonicum lipid A.
DISCUSSION
Here we describe unusual hopanoid-containing lipid A samples isolated from LPS of various strains of Bradyrhizobium. These lipid A samples had a similar sugar backbone, but differed in the number of ester-linked VLCFAs forming acyloxyacyl moieties. The identified VLCFAs had different acyl chain lengths. Additionally, all studied lipid A preparations contained at least one residue of carboxybacteriohopanediol. The substituent was ester-linked to the hydroxyl group of VLCFA, as a tertiary residue. It had been described earlier that bacteria from the Bradyrhizobium genus are able to produce a set of triterpenoids of the hopane series, as well as gammacerane derivatives. Both classes of lipids are also characteristic for R. palustris, which is closely related based on 16 S rRNA analysis and clusters together in the phylogenetic tree of the α-subgroup of proteobacteria (40). Disregard the fact that hopanoid lipids are widely distributed in the Bacteria domain, these components seem to be characteristic only of the slow-growing rhizobia. The other diazotrophic bacteria, which can fix nitrogen in endosymbiotic conditions, e.g. Rhizobium, do not contain detectable amounts of triterpenoid lipids (28, 41). Diploptene, diplopterol, 2β-methyldiplopterol, aminobacteriohopanetriol, and adenosylhopane, accompanied by tetrahymanol and their methylated homologues: 2β-methyltetrahymanol, 20α-methyltetrahymanol, and 2β,20α-dimethyltetrahymanol, were identified in cells of B. japonicum USDA 110 (28, 29). The hopanoid content in bradyrhizobial cells is usually high, and in some strains accounts for more than 40% of the total lipid fraction (28). Apart of their presence in bradyrhizobial cells, the 2-methylbacteriohopanepolyols are frequently found in cyanobacteria and are even proposed to be a biomarker of these prokaryotes (41, 42).
Very recently, the first hopanoid-containing lipid A, obtained from LPS of the photosynthetic Bradyrhizobium strain BTAi1, was structurally and functionally characterized (32). In this lipid A, one of the two ester-linked fatty acids was further substituted by one hopanoid residue.
In our work we identified two hopanoid lipids, i.e. 34-carboxyl-bacteriohopane-32,33-diol and its methylated derivative 34-carboxyl-2β-methyl-bacteriohopane-32,33-diol. They were present in LPS preparations obtained from B. japonicum, B. yuanmingense, and Bradyrhizobium sp. (Lupinus) strains. MS and NMR data proved that these hopanoid residues were covalently linked to the lipid A, forming a tertiary acyl substituent. The hydrophobic character of hopanoid rings together with the presence of two hydroxyl groups in the side chain, determined their particular position in the membrane. The VLCFAs, which were found to be components of all bradyrhizobial lipid A samples, contained between 26 and 34 carbon atoms, and were present in mono- and dimethyl branched and straight forms. These fatty acids were parts of two or, in case of B. yuanmingense lipid A, even three acyloxyacyl residues and may span the entire outer membrane. When one or two are additionally substituted by a hopanoid residue, their hydroxy groups could be placed at the boundary of the double leaflet, whereas the hydrophobic rings could be hidden inside the membrane. Thus, the presence of such unusual lipid A substituents is thought to have a strong influence on the membrane properties and act as a stabilizer of the membrane bilayer. However, additional studies are needed to establish the biophysical properties of such macromolecules and enlighten their possible function in the bacterial outer membrane. In case of lipid A from the photosynthetic Bradyrhizobium strain it was proven, by biophysical analysis of reconstituted asymmetric liposomes, that the architecture of this unusual lipid A was optimally suited to induce a high ordering of the outer membrane, reinforcing its stability and rigidity (32). Furthermore, hopanoid lipids of nitrogen-fixing bacteria (Frankia) are proposed to form a kind of diffusion barrier to protect the oxygen-sensitive nitrogrenase-hydrogenase complex from oxidative damage (27). This may also hold true for Bradyrhizobium, which, in contrast to Rhizobium, are able to fix nitrogen also in the free-living state (non-symbiotically). Our studies proved that the lipid A backbone of LPS from all examined strains were composed of a d-GlcpN3N-disaccharide, substituted at position C-4′ by an α-d-Manp-(1→6)-α-d-Manp disaccharide, whereas the position C-1 was occupied by α-(1→1)-linked d-GalpA.
The presence of d-GlcpN3N in the lipid A backbone of the LPS of nitrogen-fixing bacteria is rather common. This amino sugar was reported for lipid A of the LPS from Mesorhizobium loti (18, 43), M. huakuii (20), A. caulinodans (24), and other symbiotic bacteria belonging to the genera Ochrobactrum and Phyllobacterium.3 d-GlcpN3N was also found in lipid A derived from other, non-rhizobial bacteria, e.g. Rhodopseudomonas (where the presence of this amino sugar was described for the first time) (44), Thiobacillus sp. (45), pathogenic Brucella abortus (46), and Campylobacter jejuni (47), and also in the hyperthermophilic bacterium Aquifex pyrophilus (48).
Mannose-containing lipid A samples were identified earlier in the predatory bacterium Bdellovibrio bacteriovorus, where mannose residues occupied positions C-1 and C-4′ of the d-GlcpN3N-disaccharide (49), and in phototrophic bacterium Rhodomicrobium vannielli (50), in which the C-4′ of the glucosaminyl disaccharide backbone was occupied by one mannose residue. Recently, we reported the presence of a neutral mannose-containing lipid A in LPS of B. elkanii USDA 76 (21). In this bacterium it was demonstrated that two mannose residues forming a disaccharide were linked to C-4′ and one residue to C-1 of the d-GlcpN3N-disaccharide.
In B. japonicum USDA 110 position C-1 of the lipid A backbone was substituted by an α-(1→1)-linked d-GalpA. This unique substitution of the lipid A backbone had been noticed earlier in Mesorhizobium huakuii (20) and Mesorhizobium loti strains (43), which contained a non-reducing trisaccharide backbone comprising a β-(1→6)-d-GlcpN3N disaccharide partly substituted with phosphate at C-4′ and an α-(1→1)-linked d-GalpA in stoichiometric amounts. Other microorganisms that produce lipid A substituted with α-(1→1)-d-GalpA include A. pyrophilus (48), the stalk-forming bacterium Caulobacter crescentus (51), and the plant-growth promoting bacterium Azospirillum lipoferum (52). In the last case, the lipid A backbone constituted a GlcpN-disaccharide. In Caulobacter and Aquifex the β-(1→6)-linked d-GlcpN3N disaccharide was substituted with d-GalpA at C-4′ and C-1.
The genes responsible for this unusual glycosylation of lipid A backbone have been identified recently. A gene cluster that encoded for d-GalA transferase (rgtF, ArnT-like glycosyl transferase), lipid A 1-phosphatase (lpxE), and glycosyl transferase (rgtE), in the relative order: rgtE, duf, rgtF, and lpxE (where duf is a domain with unknown function), was identified in M. loti MAFF303099 genome (43). Similar gene clusters were found also in other mesorhizobial strains (Mesorhizobium ciceri WSM1271 and Mesorhizobium opportunistum WSM2075), as well as in C. crescentus CB15 and A. lipoferum B510 (43). BLAST similarity searches of B. japonicum USDA 110 genome revealed the presence of a similar gene cluster, which contained putative rgtF, lpxE, and rgtE genes, encoding proteins with high sequence identity (37–55%) with respective proteins of M. loti (data not shown), and thus proving the presence of genes necessary for the synthesis of α-(1→1)-d-GalpA substituted lipid A in this strain. Brown and co-workers showed that the R. leguminosarum rgtE− mutant, which produces LPS completely devoid of d-GalpA, was severely compromised in membrane stability (43). Thus, we assume that the synthesis of d-GalpA-containing lipid A by Bradyrhizobium bacteria, apart from the presence of the hopanoid substituent, may cause an additional stabilization of the outer membrane, most probably by the presence of divalent cationic bridging, which is similar to mammalian pathogens that produce phosphate-containing LPS (lipid A) (10, 43).
Acknowledgments
We are very grateful to Regina Engel and Hermann Moll for GC-MS analyses of hopanoids, Brigitte Kunz and Helga Lüthje for mass spectrometry analyses, and Heiko Käßner for recording NMR spectra (all from Research Center Borstel, Germany).
This work was supported by Polish Ministry of Science and Higher Education Grants 303 109 32/3593 and N N303 822840 (to A. Ch. and I. K.).
A. Choma, personal communication.
- VLCFA
- very long chain (ω-1)-hydroxy fatty acids
- COSY
- 1H/1H correlation spectroscopy
- DQF-COSY
- 1H/1H double quantum filtered correlation spectroscopy
- d-GlcpN
- d-glucosamine
- d-GlcpN3N
- 2,3-dideoxy-2,3-diamino-d-glucose
- ESI
- electrospray ionization
- FT-ICR MS
- Fourier-transform ion cyclotron resonance mass spectrometry
- HMBC
- 1H/13C heteronuclear multiple quantum correlation
- HSQC-DEPT
- 1H/13C heteronuclear single quantum coherence-distortionless enhancement by polarization transfer
- HSQCnd
- non-decoupled HSQC spectrum
- ROESY
- rotating frame nuclear Overhauser effect spectroscopy
- TLR4-MD-2
- Toll-like receptor 4 and myeloid differentiation factor 2 complex
- TOCSY
- 1H/1H total correlation spectroscopy.
REFERENCES
- 1. Komaniecka I., Zdzisinska B., Kandefer-Szerszen M., Choma A. (2010) Endotoxic activity of lipopolysaccharides isolated from Bradyrhizobium, Mesorhizobium and Azospirillum strains. Microbiol. Immunol. 54, 717–725 [DOI] [PubMed] [Google Scholar]
- 2. de Maagd R. A., Rao A. S., Mulders I. H., Goosen-de Roo L., van Loosdrecht M. C., Wijffelman C. A., Lugtenberg B. J. (1989) Isolation and characterization of mutants of Rhizobium leguminosarum bv. viciae 248 with altered lipopolysaccharides: possible role of surface charge or hydrophobicity in bacterial release from the infection thread. J. Bacteriol. 171, 1143–1150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brewin N. J., Perotto S., Kannenberg E. L., Rae A. L., Rathbun E. A., Lucas M. M., Kardailsky I., Gunder A., Bolanos L., Donovan N., Drobak B. K., Nester E. W., Verma D. P. (1993) Mechanisms of cell and tissue invasion by Rhizobium leguminosarum: the role of cell surface interactions. in Current Plant Science and Biotechnology in Agriculture, Advances in Molecular Genetics of Plant-Microbe Interactions (Nestery E. W., Verma D. P. S., eds) Vol. 2, pp. 369–380, Kluwer Academic Publishers, Dordrecht/Boston/London [Google Scholar]
- 4. Fraysse N., Jabbouri S., Treilhou M., Couderc F., Poinsot V. (2002) Symbiotic conditions induce structural modifications of Sinorhizobium sp. NGR234 surface polysaccharides. Glycobiology 12, 741–748 [DOI] [PubMed] [Google Scholar]
- 5. Carlson R. W., Forsberg L. S., Kannenberg E. L. (2010) Lipopolysaccharides in Rhizobium-legume symbioses. in Endotoxins: structure, function and recognition (Wang X., Quinn P. J., eds) pp. 339–386, Springer, The Netherlands: [DOI] [PubMed] [Google Scholar]
- 6. Dow M., Newman M. A., von Roepenack E. (2000) The induction and modulation of plant defense responses by bacterial lipopolysaccharides. Annu. Rev. Phytopathol. 38, 241–261 [DOI] [PubMed] [Google Scholar]
- 7. Albus U., Baier R., Holst O., Pühler A., Niehaus K. (2001) Suppression of an elicitor-induced oxidative burst in Medicago sativa cell cultures by Sinorhizobium meliloti lipopolysaccharides. New Phytologist 151, 597–606 [DOI] [PubMed] [Google Scholar]
- 8. Mathis R., Van Gijsegem F., De Rycke R., D'Haeze W., Van Maelsaeke E., Anthonio E., Van Montagu M., Holsters M., Vereecke D. (2005) Lipopolysaccharides as a communication signal for progression of legume endosymbiosis. Proc. Natl. Acad. Sci. U.S.A. 102, 2655–2660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zähringer U., Lindner B., Rietschel E. Th. (1999) Chemical structure of lipid A: recent advances in structural analysis of biologically active molecules. in Endotoxin in Health and Disease (Brade H., Opal S. M., Vogel S. N., Morrison D. C., eds) pp. 93–114, Marcel Dekker, New York [Google Scholar]
- 10. Raetz C. R., Whitfield C. (2002) Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 71, 635–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Park B. S., Song D. H., Kim H. M., Choi B.-S., Lee H., Lee J.-O. (2009) The structural basis of lipopolysaccharide recognition by the TLR4–MD-2 complex. Nature 458, 1191–1195 [DOI] [PubMed] [Google Scholar]
- 12. Beutler B., Rietschel E. T. (2003) Innate immune sensing and its roots: the story of endotoxin. Nat. Rev. Immunol. 3, 169–176 [DOI] [PubMed] [Google Scholar]
- 13. Poltorak A., He X., Smirnova I., Liu M. Y., Van Huffel C., Du X., Birdwell D., Alejos E., Silva M., Galanos C., Freudenberg M., Ricciardi-Castagnoli P., Layton B., Beutler B. (1998) Defective LPS Signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science 282, 2085–2088 [DOI] [PubMed] [Google Scholar]
- 14. Urbanik-Sypniewska T., Choma A., Kutkowska J., Kamińska T., Kandefer-Szerszeń M., Russa R., Dolecka J. (2000) Cytokine inducing activities of rhizobial and mesorhizobial lipopolysaccharides of different lethal toxicity. Immunobiology 202, 408–420 [DOI] [PubMed] [Google Scholar]
- 15. Tsukushi Y., Kido N., Saeki K., Sugiyama T., Koide N., Mori I., Yoshida T., Yokochi T. (2004) Characteristic biological activities of lipopolysaccharides from Sinorhizobium and Mesorhizobium. J. Endotoxin Res. 10, 25–31 [DOI] [PubMed] [Google Scholar]
- 16. Bhat U. R., Forsberg L. S., Carlson R. W. (1994) Structure of lipid A component of Rhizobium leguminosarum bv. phaseoli lipopolysaccharide. J. Biol. Chem. 269, 14402–14410 [PubMed] [Google Scholar]
- 17. Gil-Serrano A. M., González-Jiménez I., Tejero-Mateo P., Megías M., Romero-Vazquez M. J. (1994) Analysis of the lipid moiety of lipopolysaccharide from Rhizobium tropici CIAT899: identification of 29-hydroxytriacontanoic acid. J. Bacteriol. 176, 2454–2457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Russa R., Urbanik-Sypniewska T., Lindström K., Mayer H. (1995) Chemical characterization of two lipopolysaccharide species isolated from Rhizobium loti NZP2213. Arch. Microbiol. 163, 345–351 [DOI] [PubMed] [Google Scholar]
- 19. Que N. L., Lin S., Cotter R. J., Raetz C. R. (2000) Purification and mass spectrometry of six lipid A species from the bacterial endosymbiont Rhizobium etli. J. Biol. Chem. 275, 28006–28016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Choma A., Sowinski P. (2004) Characterization of Mesorhizobium huakuii lipid A containing both d-galacturonic acid and phosphate residues. Eur. J. Biochem. 271, 1310–1322 [DOI] [PubMed] [Google Scholar]
- 21. Komaniecka I., Choma A., Lindner B., Holst O. (2010) The structure of a novel neutral lipid A from the lipopolysaccharide of Bradyrhizobium elkanii containing three mannoses units in the backbone. Chem.-Eur. J. 16, 2922–2929 [DOI] [PubMed] [Google Scholar]
- 22. Bhat U. R., Carlson R.W., Busch M., Mayer H. (1991) Distribution and phylogenetic significance of 27-hydroxy-octacosanoic acid in lipopolysaccharides from bacteria belonging to the α-2 subgroup of Proteobacteria. Int. J. Syst. Bacteriol. 41, 213–217 [DOI] [PubMed] [Google Scholar]
- 23. Bhat U. R., Mayer H., Yokota A., Hollingsworth R. I., Carlson R. W. (1991) Occurrence of lipid A variants with 27-hydroxyoctacosanoic acid in lipopolysaccharides from members of the family Rhizobiaceae. J. Bacteriol. 173, 2155–2159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Choma A., Komaniecka I., Turska-Szewczuk A., Danikiewicz W., Spolnik G. (2012) Structure of lipid A from a stem-nodulating bacterium Azorhizobium caulinodans. Carbohydr. Res. 352, 126–136 [DOI] [PubMed] [Google Scholar]
- 25. Choma A., Komaniecka I. (2011) Straight and branched ω-1 hydroxylated very long chain fatty acids are components of Bradyrhizobium lipid A. Acta Biochim. Pol. 58, 51–58 [PubMed] [Google Scholar]
- 26. Neunlist S., Holst O., Rohmer M. (1985) Prokaryotic triterpenoids. The hopanoids of the purple non-sulphur bacterium Rhodomicrobium vannielii: an aminotriol and its aminoacyl derivatives, N-tryptophanyl and N-ornithinyl aminotriol. Eur. J. Biochem. 15, 561–568 [PubMed] [Google Scholar]
- 27. Berry A. M., Harriott O. T., Moreau R.A., Osman S. F., Benson D. R., Jones A. D. (1993) Hopanoid lipids compose the Frankia vesicle envelope, presumptive barrier of oxygen diffusion to nitrogenase. Proc. Natl. Acad. Sci. U.S.A. 90, 6091–6094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kannenberg E. L., Perzl M., Härtner T. (1995) The occurrence of hopanoid lipids in Bradyrhizobium bacteria. FEMS Microbiol. Lett. 127, 255–262 [DOI] [PubMed] [Google Scholar]
- 29. Bravo J.-M., Perzl M., Härtner T., Kannenberg E. L., Rohmer M. (2001) Novel methylated triterpenoids of the gammacerane series from the nitrogen-fixing bacterium Bradyrhizobium japonicum USDA 110. Eur. J. Biochem. 268, 1323–1331 [DOI] [PubMed] [Google Scholar]
- 30. Welander P. V., Hunter R. C., Zhang L., Sessions A. L., Summons R. E., Newman D. K. (2009) Hopanoids play a role in membrane integrity and pH homeostasis in Rhodopseudomonas palustris TIE-1. J. Bacteriol. 191, 6145–6156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rohmer M., Bouvier P., Ourisson G. (1979) Molecular evolution of biomembranes: structural equivalents and phylogenetic precursors of sterols. Proc. Natl. Acad. Sci. U.S.A. 76, 847–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Silipo A., Vitiello G., Gully D., Sturiale L., Chaintreuil C., Fardoux J., Gargani D., Lee H. I., Kulkarni G., Busset N., Marchetti R., Palmigiano A., Moll H., Engel R., Lanzetta R., Paduano L., Parrilli M., Chang W. S., Holst O., Newman D. K., Garozzo D., D'Errico G., Giraud E., Molinaro A. (2014) Covalently linked hopanoid-lipid A improves outer-membrane resistance of a Bradyrhizobium symbiont of legumes. Nat. Commun. 5, 5106, 10.1038/ncomms6106 [DOI] [PubMed] [Google Scholar]
- 33. Vincent M. (1970) A manual for the practical study of root-nodule bacteria. International Biological Programme, Handbook No. 15. Blackwell, Oxford [Google Scholar]
- 34. Reitz M., Rudolph K., Schröder I., Hoffmann-Hergarten S., Hallmann J., Sikora R. A. (2000) Lipopolysaccharides of Rhizobium etli strain G12 act in potato roots as an inducing agent of systemic resistance to infection by the cyst nematode Globodera pallida. Appl. Environ. Microbiol. 66, 3515–3518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Westphal O., Jann K. (1965) Bacterial lipopolysaccharide: extraction with phenol-water and further application of the procedure. in Methods in Carbohydrate Chemistry (Whistler R. L., ed) Vol. 5, pp. 83–91, Academic Press, New York [Google Scholar]
- 36. Johnson K. G., Perry M. B. (1976) Improved techniques for the preparation of bacterial lipopolysaccharides. Can. J. Microbiol. 22, 29–34 [DOI] [PubMed] [Google Scholar]
- 37. Que-Gewirth N. L., Ribeiro A. A., Kalb S. R., Cotter R. J., Bulach D. M., Adler B., Girons I. S., Werts C., Raetz C. R. (2004) A methylated phosphate group and four amide-linked acyl chains in Leptospira interrogans lipid A. J. Biol. Chem. 279, 25420–25429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Simonin P., Tindall B., Rohmer M. (1994) Structure elucidation and biosynthesis of 31-methylhopanoids from Acetobacter europaeus. Eur. J. Biochem. 225, 765–771 [DOI] [PubMed] [Google Scholar]
- 39. Simonin P., Jürgens U. J., Rohmer M. (1996) Bacterial triterpenoids of the hopane series from the Prochlorothrix hollandica and their intracellular localization. Eur. J. Biochem. 241, 865–871 [DOI] [PubMed] [Google Scholar]
- 40. Young J. P., Downer H. L., Eardly B. D. (1991) Phylogeny of the phototrophic Rhizobium strain BTAi1 by polymerase chain reaction-based sequencing of a 16 S rRNA gene segment. J. Bacteriol. 173, 2271–2277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rohmer M., Bouvier-Nave P., Ourisson G. (1984) Distribution of hopanoid triterpenes in prokaryotes. J. Gen. Microbiol. 130, 1137–1150 [Google Scholar]
- 42. Summons R. E., Jahnke L. L., Hope J. M., Logan G. A. (1999) 2-Methylhopanoids as biomarkers for cyanobacterial oxygenic photosynthesis. Nature 400, 554–557 [DOI] [PubMed] [Google Scholar]
- 43. Brown D. B., Muszynski A., Carlson R. W. (2013) Elucidation of a novel lipid A α-(1,1)-GalA transferase gene (rgtF) from Mesorhizobium loti: heterologous expression of rgtF causes Rhizobium etli to synthesize lipid A with α-(1,1)-GalA. Glycobiology 23, 546–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Roppel J., Mayer H. (1975) Identification of a 2,3-diamino-2,3-dideoxyhexose in the lipid A component of lipopolysaccharides of Rhodopseudomonas viridis and Rhodopseudomonas palustris. Carbohydr. Res. 40, 31–40 [DOI] [PubMed] [Google Scholar]
- 45. Yokota A., Rodriguez M., Yamada Y., Imai K., Borowiak D., Mayer H. (1987) Lipopolysaccharides of Thiobacillus species containing lipid A with 2,3-diamino-2,3-dideoxyglucose. Arch. Microbiol. 149, 106–111 [Google Scholar]
- 46. Moreno E., Stackebrandt E., Dorsch M., Wolters J., Busch M., Mayer H. (1990) Brucella abortus 16S rRNA and lipid A reveal a phylogenetic relationship with members of the α-2 subdivision of the class Proteobacteria. J. Bacteriol. 172, 3569–3576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Moran A. P., Zähringer U., Seydel U., Scholz D., Stütz P., Rietschel E. T. (1991) Structural analysis of lipid A component of Campylobacter jejuni CCUG 10936 (serotype O:2) lipopolysaccharide. Eur. J. Biochem. 198, 459–469 [DOI] [PubMed] [Google Scholar]
- 48. Plötz B. M., Lindner B., Stetter K. O., Holst O. (2000) Characterization of a novel lipid A containing d-galacturonic acid that replaces phosphate residues. J. Biol. Chem. 275, 11222–11228 [DOI] [PubMed] [Google Scholar]
- 49. Schwudke D., Linscheid M., Strauch E., Appel B., Zahringer U., Moll H., Muller M., Brecker L., Gronow S., Lindner B. (2003) The obligate predatory Bdellovibrio bacteriovorus possesses a neutral lipid A containing α-d-mannoses that replace phosphate residues. J. Biol. Chem. 278, 27502–27512 [DOI] [PubMed] [Google Scholar]
- 50. Holst O., Borowiak D., Weckesser J., Mayer H. (1983) Structural studies on the phosphate-free lipid A of Rhodomicrobium vannielii ATCC 17100. Eur. J. Biochem. 137, 325–332 [DOI] [PubMed] [Google Scholar]
- 51. Smit J., Kaltashov I. A., Cotter R. J., Vinogradov E., Perry M. B., Haider H., Qureshi N. (2008) Structure of a novel lipid A obtained from the lipopolysaccharide of Caulobacter crescentus. Innate Immun. 14, 25–37 [DOI] [PubMed] [Google Scholar]
- 52. Choma A., Komaniecka I. (2008) Characterization of a novel lipid A structure isolated from Azospirillum lipoferum lipopolysaccharide. Carbohydr. Res. 343, 799–804 [DOI] [PubMed] [Google Scholar]