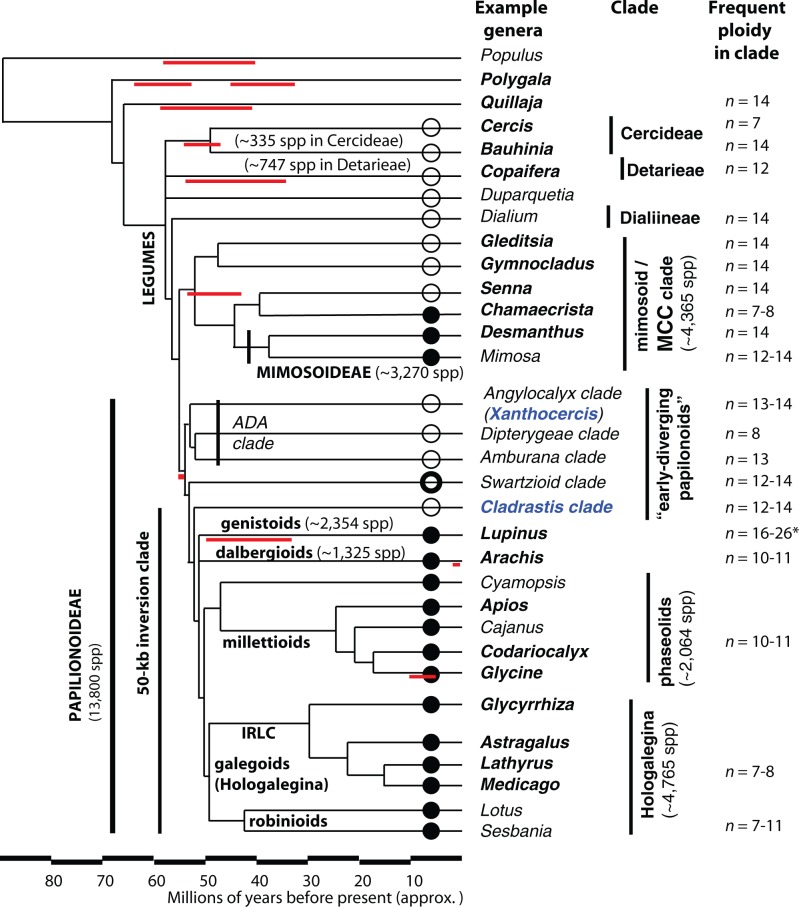
Fig. 1.
A summary phylogeny based on published trees for the Fabaceae (Wojciechowski et al. 2004; Cardoso et al. 2012; Manzanilla and Bruneau 2012), with approximate date estimates from Lavin et al. (2005). Estimates of species counts per clade are taken from Lewis et al. (2005). Nodulation status is shown with circles: Filled for “many species in this clade nodulate”; partly filled (Swartzieae) for “some species in this clade nodulate”; empty for “no species in this clade have been observed to nodulate.” Nodulation status is summarized from Sprent (2009). Chromosome counts on the right are predominant counts for each genus or clade. These are drawn from Doyle (2012) and supplementary file S5, Supplementary Material online. For the genistoids, the base chromosome count is likely x = 9, but chromosome counts within Lupinus (used in this project) are mostly in the range of n = 16–26 (Doyle 2012). Hypothesized placement of genome duplication events is indicated with horizontal red lines.