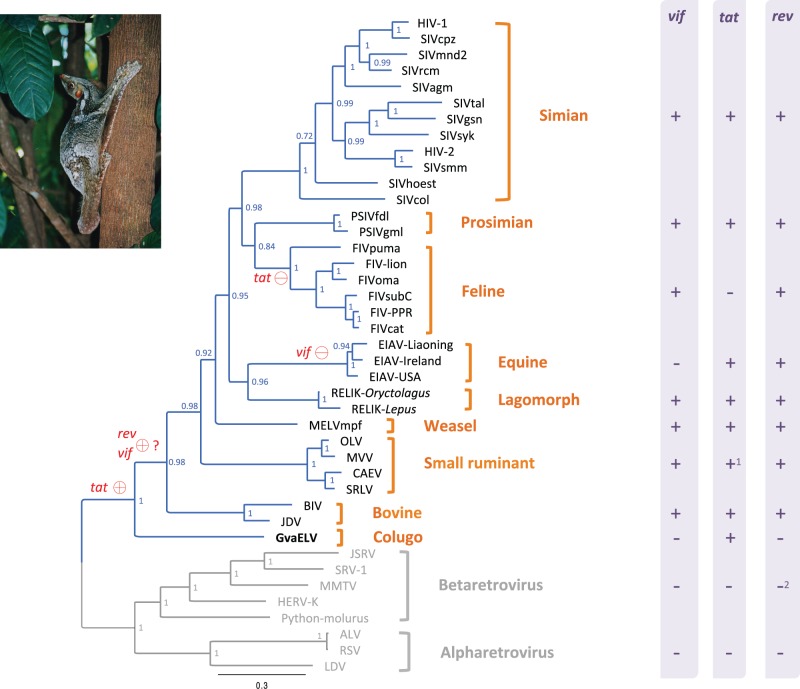
Fig. 2.
Phylogeny and genomic structure diversity among exogenous and endogenous lentiviruses. The phylogeny was reconstructed based on the concatenated sequences of Gag and Pol proteins. The phylogeny is a 50% majority-rule consensus tree. Node labels are posterior probabilities. The lentivirus lineage is highlighted in blue. The presence and absence of accessory genes (vif, tat, and rev) in lentiviruses, alpharetroviruses, and betaretroviruses are indicated by + and − in the right column, respectively. Red circles on branches indicate the most parsimonious scenario of birth (+) and death (−) of related accessory genes. Note 1: Small ruminant lentivirus (SLRV) encodes a tat ORF that shares discernible similarity with tat gene of other lentiviruses, although the SLRV Tat protein is not functionally similar to that of other lentiviruses (Villet et al. 2003). Note 2: Although some betaretroviruses, such as mouse mammary tumor virus and human ERV K, encode functional homologs of rev genes (Yang et al. 1999; Mertz et al. 2005; Gifford 2012), these betaretrovirus proteins do not share discernible sequence similarity with lentiviral Rev protein (Yang et al. 1999). Virus names: ALV, avian leukemia virus; BIV, bovine immunodeficiency virus; CAEV, caprine arthritis–encephalitis virus; HERV-K, human ERV K; HIV, human immunodeficiency virus; JDV, Jembrana disease virus; JSRV, jaagsiekte sheep retrovirus; LDV, lymphoproliferative disease virus; MMTV, mouse mammary tumor virus; MVV, maedi-visna virus; OLV, olive lentivirus; Python-molurus, Python molurus ERV; RSV, Rous sarcoma virus; SRLV, small ruminant lentivirus; SRV-1, Simian retrovirus 1. Colugo photo courtesy of Nina Holopainen.