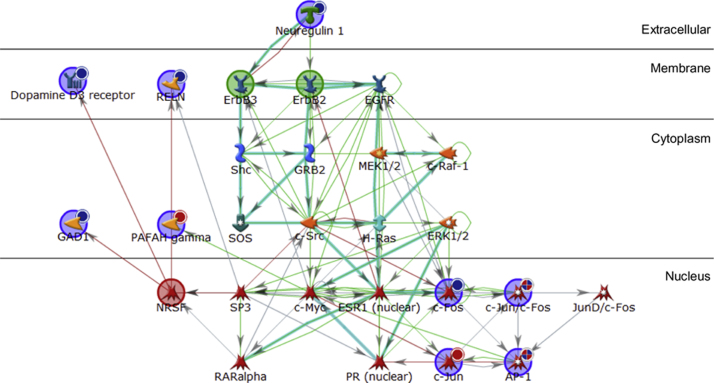
Fig. 1.
Network analysis of genes significantly modulated in response to mood stabilisers. Genes shown to be significantly up or down regulated in human SH-SY5Y cells in response to 1 h treatment with the mood stabilisers sodium valproate and lithium were uploaded into MetaCore™ for network analysis. The gene list was analysed under the Build Network feature using the Transcription Factor Targets Modelling algorithm. Seed nodes from which the network was built upon are encompassed by a large circle; blue circles represent genes from the experimental data, green circles represent molecules from which the pathway is expanded from and red circles represent molecules on which the pathway terminates. Genes uploaded from the experimental data are also marked with a smaller circle in their top right hand corner; red circles represent genes that were significantly up-regulated, whereas blue circles represent genes significantly down-regulated. Connecting arrows indicate interactions; green arrows represent activation, red arrows represent inhibition and blue arrows are unspecified. Overlaid cyan lines represent canonical pathways. Gene names/symbols within the network from top to bottom, left to right: Neuregulin 1, Dopamine D3 receptor, RELN, ErbB3, ErbB2, EGFR, Shc, GRB2, MEK1/2, c-Raf-1, GAD1 PAFAH gamma, SOS, c-Src, H-Ras, ERK1/2, NRSF, SP3, c-Myc, ESR1 (nuclear), c-Fos, c-Jun/c-Fos, JunD/c-Fos, RARalpha, PR (nuclear) c-Jun, and AP-1. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)