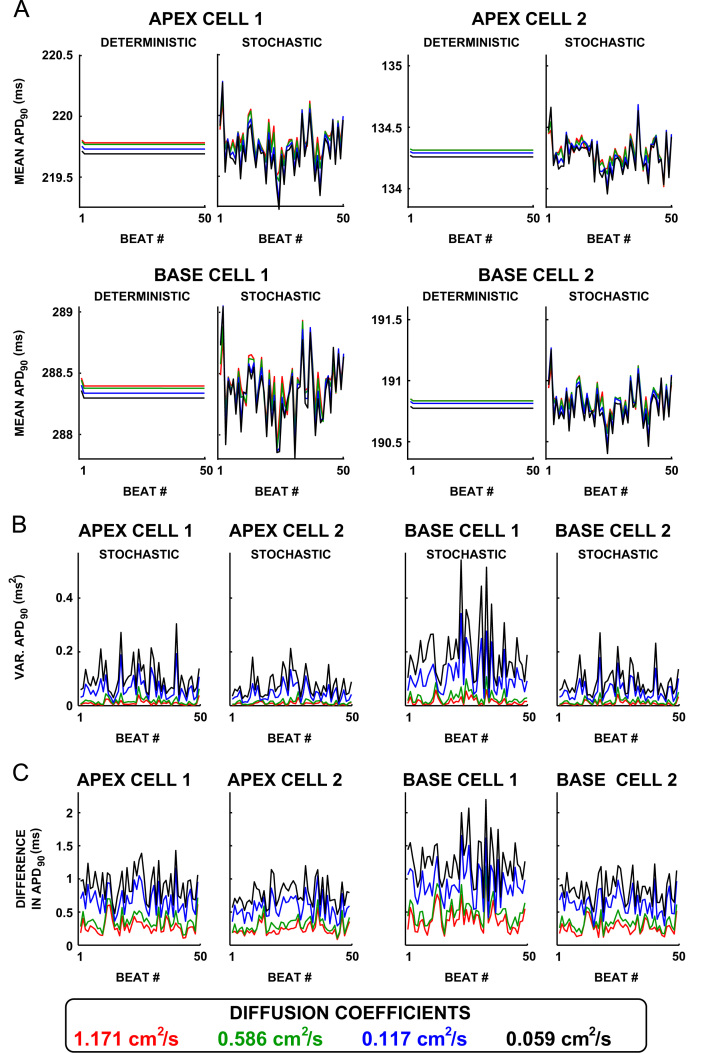
Fig. 2.
Reducing coupling in tissue simulations with intrinsic variability increases dispersion in APD90. (A) The mean APD90 across all nodes in the central 0.5 cm of the tissue is plotted for both the deterministic (left) and stochastic (right) simulations for each parameter set and each value of D. The values of D shown are 1.171 cm2 s−1 (red), D=0.586 cm2 s−1 (green), D= 0.117 cm2 s−1 (blue), and D= 0.059 cm2 s−1 (black). (B) The difference between the stochastic and deterministic APD90 maps is calculated at each node and each AP in the central 0.5 cm of the tissue. The variance across these nodes is then calculated for each beat. The variance in the difference between the deterministic and stochastic APD90 maps is plotted as a function of the number of beats. (C) The absolute difference between the deterministic and stochastic simulations is calculated at each node in the central 0.5 cm of the tissue. The maximum of these absolute differences for each beat are then plotted as a function of the number of beats. (For interpretation of the references to colour in this figure caption, the reader is referred to the web version of this paper.)