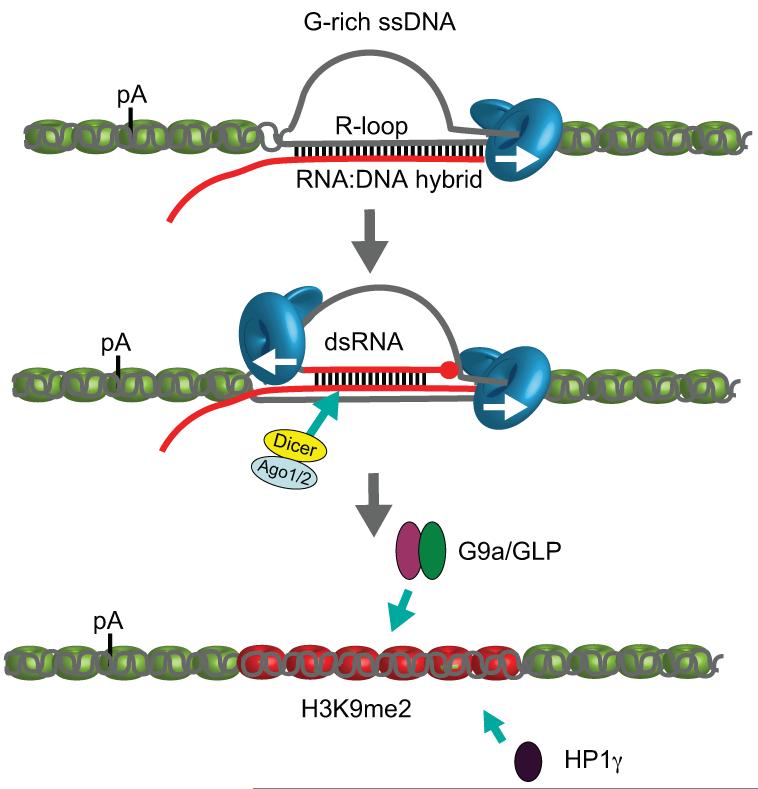
ED Figure 10. Model for how R-loops and RNAi-dependent H3K9me2 chromatin mediate pause-dependent transcriptional termination in mammalian genes.
Mammalian genes possessing pause elements downstream of their PAS form R-loops in termination regions. This facilitates generation of an antisense transcript that hybridises with the sense transcript to form dsRNA. This triggers recruitment of the RNAi factors, Dicer, Ago1 and Ago2. G9a/GLP HKMTs and HP1γ are then recruited forming and maintaining H3K9me2 repressive marks. R-loops and H3K9me2 facilitate Pol II pausing prior to termination. DNA is shown as grey lines and RNA as a red line. Points of contact between the DNA strand and nascent RNA indicates R-loop formation, whereas points of contact between sense and antisense RNA indicate dsRNA formation. Pol II is shown as a blue icon with arrow indicating transcription direction. Nucleosomes are shown in green except over H3K9me2 region where they are coloured red.