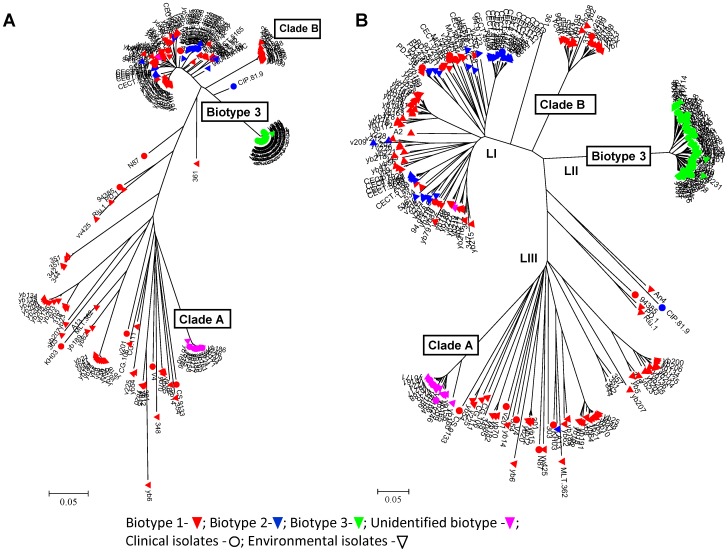
Figure 2. Phylogenetic relationships among 254 V. vulnificus isolates based on variation data at 570 SNP loci.
(A) Parametric analysis: the evolutionary history was inferred using the neighbor-joining method and the evolutionary distances were computed using the Kimura two-parameter method with bootstrap analysis of 1,000 replicates. (B) Nonparametric analysis: genetic relationships among strains were inferred using the Jaccard similarity coefficient, and the distance matrix was generated with PAST software (version 1.94b). The phylogenetic tree was constructed by using the minimum evolution (ME) cluster analysis.