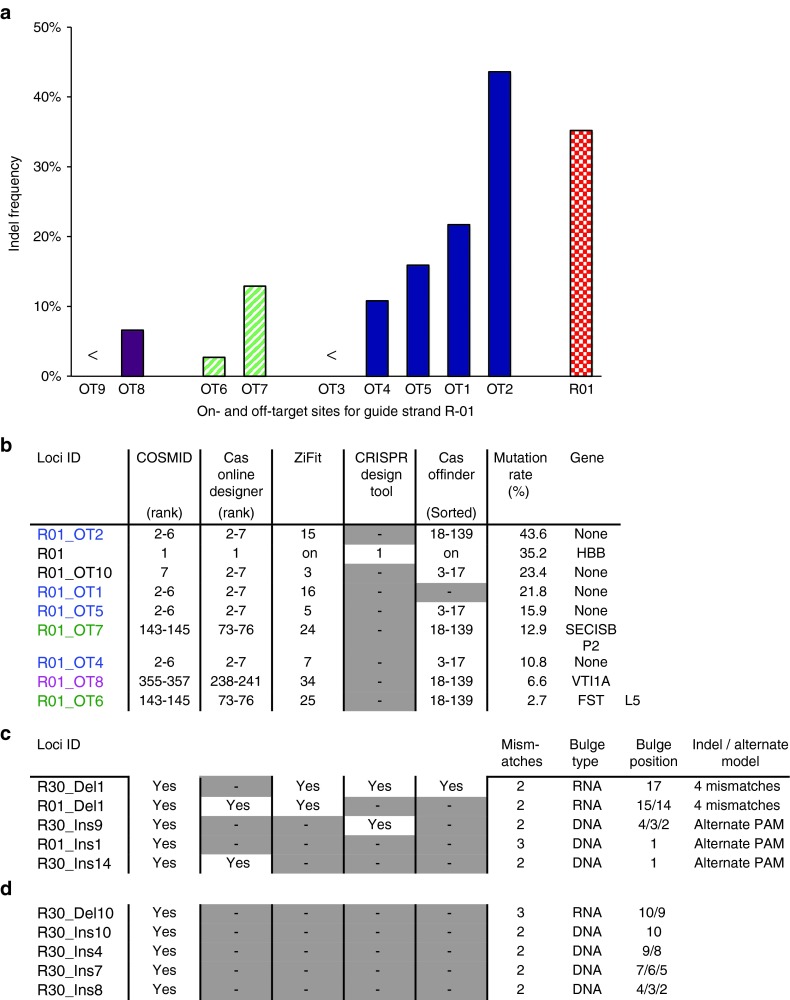
Figure 2.
Comparison of COSMID with other available tools in predicting off-target sites for guide strand R-01. (a) On- and off-target cleavage rates for guide strand R-01. Marked differences are seen in the cleavage rates at off-target sites with identical sequences, but different chromosomal locations, such as OT8 and OT9 (purple), OT6 and OT7 (green stripes), and OT1–OT5 (blue). The R-01 on-target indel rate is shown to the right (red pattern). (b–d) Comparison of COSMID with other available tools in predicting off-target sites for guide strand R-01. (b) Comparison of RO1 off-target sites that contain two mismatches. The cleavage rates at R-01 on-target site and off-target sites OT1–OT10 are listed by decreasing T7EI activity. OT3 and OT9 had activities below T7E1 detection limit. Sites with matching sequences (outside first base) have their names in bold with colors matching that shown in a. Annotated genes corresponding to the sites are listed. Off-target analysis was performed with different online search tools. If the genomic sites with measurable T7E1 activity (shown in a) were identified by a specific tool (such as Cas OFFinder), their rankings in its output (if sortable) are shown. Sites not in the output of that tool are indicated by a dash in a grey box. (c) Comparison of search results for off-target sites that contain deletions or insertions, in which sequence-verified off-target sites with insertions or deletions, which can also be modeled as loci with four mismatches or alternate PAM considered. (d) The sequence-verified off-target sites with insertions or deletions that cannot be modeled as four mismatches or alternate PAM can only be predicted by COSMID.