Figure 1.
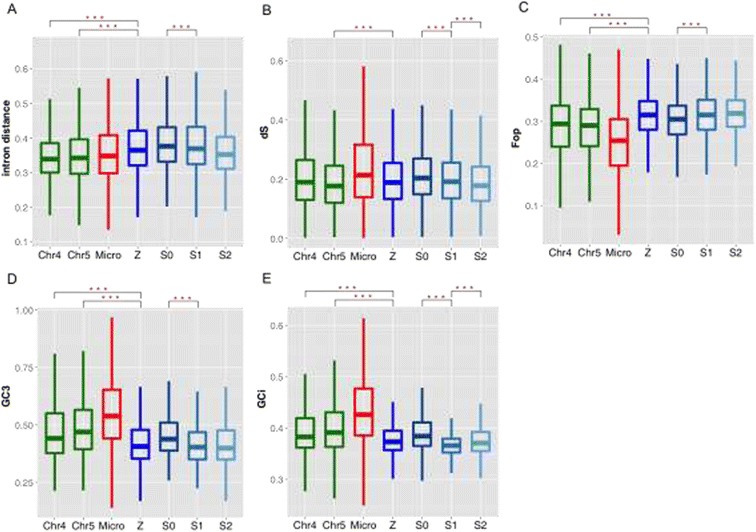
Evolution patterns of synonymous sites and non-coding regions in bird genomes. (A-E) We show sequence patterns in introns and synonymous sites of coding regions divided in different chromosome sets or different evolutionary strata (from young to old strata: S2, S1, S0) of Z chromosomes. Microchromosome genes (shown in red) exhibit a very different sequence pattern (dS, codon usage bias, G + C content) relative to macro-chromosomes including the Z, due to their different chromosome size and gene densities [50]. Therefore, we mainly compare the patterns of Z-linked genes to those of chr4 and chr5, which have a similar size and gene number. We also performed comparisons between Z chromosome vs. all macro-chromosomes, and the results are similar (Additional file 3: Table S2). We show the Wilcoxon test significance level of differences with asterisks. ***:P-value < 0.001. dS: lineage specific synonymous substitution rate. Fop: frequency of optimal codons. GC3: G + C content at the third codon position. GCi: G + C content of introns.
