Abstract
The bacterial strategy of chemotaxis relies on temporal comparisons of chemical concentrations, where the probability of maintaining the current direction of swimming is modulated by changes in stimulation experienced during the recent past. A short-term memory required for such comparisons is provided by the adaptation system, which operates through the activity-dependent methylation of chemotaxis receptors. Previous theoretical studies have suggested that efficient navigation in gradients requires a well-defined adaptation rate, because the memory time scale needs to match the duration of straight runs made by bacteria. Here we demonstrate that the chemotaxis pathway of Escherichia coli does indeed exhibit a universal relation between the response magnitude and adaptation time which does not depend on the type of chemical ligand. Our results suggest that this alignment of adaptation rates for different ligands is achieved through cooperative interactions among chemoreceptors rather than through fine-tuning of methylation rates for individual receptors. This observation illustrates a yet-unrecognized function of receptor clustering in bacterial chemotaxis.
INTRODUCTION
In bacterial chemotaxis, effector stimuli are detected by a set of transmembrane receptors of different specificities (1–4). Escherichia coli has five types of chemoreceptors, with Tar and Tsr being major receptors and Trg, Tap, and Aer being minor chemoreceptors. Together with the histidine kinase CheA and an adaptor protein, CheW, receptors are organized in large sensory complexes that perform most of signal processing in chemotaxis (5–9). Activities of teams of 10 to 20 individual receptors are allosterically coupled within these complexes, enabling amplification and integration of changes in the relative ligand occupancy of receptors (10–17). The integrated signaling output of the receptor complexes is subsequently converted into the stimulation-dependent phosphorylation of the response regulator CheY, which controls cell swimming behavior.
The bacterial strategy of chemotaxis relies on temporal comparisons of chemoeffector concentrations. This requires a short-term memory that enables bacteria to detect changes in concentrations along the swimming track and to modify their behavior accordingly, to either continue swimming in this direction or to tumble and reorient (18, 19). Memory is provided by the receptor methylation system, consisting of the methyltransferase CheR and the methylesterase CheB, which respectively methylate or demethylate receptors on four or five specific glutamate residues. CheR preferentially recognizes the inactive state of receptors and increases receptor activity through methylation, whereas CheB preferentially demethylates active receptors and thereby lowers their activity (20–26). An additional negative feedback is provided by the CheA-mediated phosphorylation of CheB, which increases its methylesterase activity (27, 28). In the absence of a gradient, these feedbacks allow the system to adapt to ambient stimulation, ensuring that the activity of the receptor-associated kinase is adjusted to generate intermediate levels of the phosphorylated CheY (CheY-P). Since CheY-P binding to flagellar motors induces cell tumbling, such an intermediate level of CheY-P produces a random sequence of runs and tumbles that allows cells to explore their environment. Increased attractant binding to receptors inhibits CheA activity and thus CheY-P formation, so that swimming up a gradient of attractant results in suppression of tumbling and longer runs in that direction. Receptor methylation by CheR then slowly resets the kinase activity back to steady state, and this delay in adaptation functions as memory (29, 30).
Several theoretical studies suggested that optimal chemotaxis in a gradient requires a specific adaptation rate that is defined by the gradient steepness and cell swimming velocity (31–34), irrespective of the chemical nature of the ligand or of its receptor specificity. This requirement arises because the time window of temporal comparisons needs to match the typical time (a few seconds) of a straight run of a bacterial cell along the gradient. The importance of a well-defined adaptation rate for the chemotaxis strategy is emphasized by existence of specific mechanisms which ensures the robustness of this rate against variations in the levels of ambient attractant (35, 36), temperature (33) and gene expression (37, 38). This theoretical conclusion contrasts, however, with biochemical studies demonstrating that the rate of methylation depends on the type of the receptor and on the glutamyl residue (23, 39), as well as on the level of CheR (33, 37, 40, 41). This discrepancy raises questions as to whether the adaptation rates for different ligands are indeed equal (or similar), as predicted by theory, and, if so, how such a universal adaptation rate is ensured despite the receptor- and residue-specific differences in the methylation rates and variations in the levels of individual receptors.
Experimentally, the adaptation rate can be derived from measurements of the time that is needed for recovery of the pathway activity upon step-like stimulation (33, 42–44). Because more methyl groups are required to offset a stronger stimulus, the adaptation time depends both on the adaptation rate and on the strength of the initial stimulation (21, 42). In this study, we investigated the rates of adaptation to attractants sensed by different types of receptors. In wild-type cells, we observed a universal response-adaptation (RA) relation between response amplitude and adaptation time for all tested attractants, regardless of the ligand type. This invariance in the RA relation naturally emerges from allosteric interactions between receptors in sensory clusters, thus suggesting another important function for the chemoreceptor clustering observed across prokaryotes.
MATERIALS AND METHODS
Cell cultivation.
Bacterial overnight cultures were grown at 30°C in tryptone broth (TB; 1% tryptone, 0.5% NaCl) medium containing appropriate antibiotics, at final concentrations of 100 μg/ml for ampicillin and 34 μg/ml for chloramphenicol, and supplemented with inducers when needed. Expression of the CheY-YFP (yellow fluorescent protein)/CheZ-CFP (cyan fluorescent protein) fluorescence resonance energy transfer (FRET) pair from the bicistronic plasmid pVS88 was induced with 50 μM isopropyl-β-d-thiogalactopyranoside (IPTG), and receptor expression was induced by various concentrations of salicylate from pVS1086 (Tar) or pVS362 (Tsr) in the Δ(cheY cheZ) strains SN1 (wild type), VS181 (lacking all chemoreceptors), SN25 (lacking Tar), SN11 (lacking Tsr), and UU2795 (lacking Tar, Tap, Trg, and Aer) (see Table S1 in the supplemental material). For FRET experiments, these cultures were diluted 1:100 (wild type) or 1:20 (strains expressing receptor from a plasmid) into fresh medium supplemented with antibiotics and relevant inducers and grown to an optical density at 600 nm (OD600) of 0.6 at 34°C and 275 rpm. Cells were resuspended in tethering buffer (10 mM KPO4, 0.1 mM EDTA, 1 μM methionine, 10 mM sodium lactate [pH 7]) and kept at 4°C for at least 30 min prior to the measurements to terminate cell growth and protein expression (45).
FRET measurements.
Cells were attached to a polylysine-coated coverslip and kept under a constant flow of tethering buffer at a rate of 300 μl/min in a flow chamber. To add or remove attractants, the attached syringe pump was stopped briefly. Dose-response curves were measured using a FRET assay of the pathway activity (15, 17, 45) on custom-modified Zeiss AxioObserver or AxioImager microscopes by step-like stimulation of buffer-adapted cells with attractants. Cells were allowed to adapt to attractant to determine the adaptation time, defined as the time needed to regain 50% of the initial loss in the ratio of YFP to CFP fluorescence upon stimulation (Fig. 1A). The response amplitudes to individual steps in attractant concentrations were normalized to the response of buffer-adapted cells to a saturating stimulus of 100 μM α-methyl-dl-aspartate (MeAsp) or 100 μM serine, which fully inhibited the kinase activity and allowed us to determine the background YFP/CFP ratio in the absence of FRET. The relative kinase activity was calculated from the amplitudes of changes in the YFP/CFP ratio as described previously (15, 17, 33, 45). Up to a minor correction factor, relative kinase activity is directly reflected by the YFP/CFP ratio above the background value observed upon saturating stimulation.
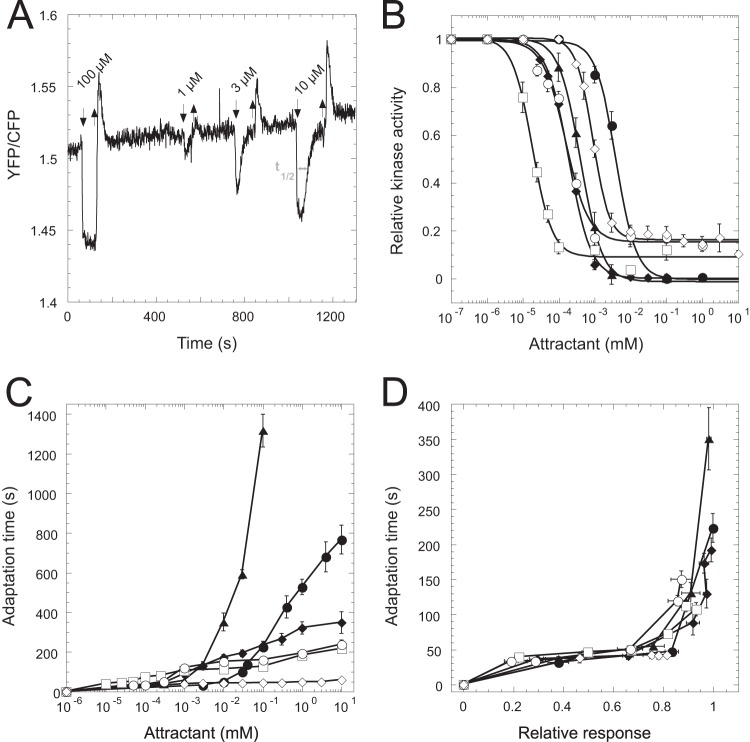
FIG 1.
Alignment of adaptation kinetics for different ligands. (A) Example FRET measurement of wild-type cell response to serine. The adaptation time (t1/2, time to recover 50% of the initial activity change) is indicated by the gray arrow. (B and C) Attractant-induced change in the relative kinase activity (B) and adaptation time (C), plotted as a function of the concentration of MeAsp (filled circles), maltose (filled diamonds), serine (triangles), galactose (squares), ribose (open circles), or Pro-Leu (open diamonds). (D) Dependence of the adaptation time on the relative response amplitude, defined as a change in the relative kinase activity. Only data points up to response saturation are shown.
RESULTS
Universal relation between chemotactic response and adaptation time.
To compare the adaptation kinetics for different chemoattractants, we measured responses and adaptation times for attractants sensed by all four types of ligand-specific E. coli chemoreceptors: α-methyl-dl-aspartate (MeAsp) and maltose for Tar, serine for Tsr, galactose and ribose for Trg, and Pro-Leu for Tap. Changes in the kinase activity upon attractant stimulation were monitored using the phosphorylation-dependent interaction between CheY-YFP and its phosphatase CheZ-CFP, detected using fluorescence resonance energy transfer (FRET) (45, 46). As expected, the response was adaptive for all attractants tested, with both the response amplitude and the time of adaptation being dependent on the chemoeffector concentration (Fig. 1A to C). More surprisingly, when the half-time of adaptation (i.e., the time to recover 50% of the initial loss in FRET signal upon stimulation) (Fig. 1A) was plotted as a function of response amplitude for the subsaturating stimuli, the dependencies for all ligands collapsed onto a single curve (Fig. 1D). This collapse demonstrates a universal relation between response amplitude and adaptation time in the chemotaxis system of E. coli in the subsaturating range of stimulus strength. Such universal response-adaptation (RA) relation suggests that the rate of adaptation depends solely on the extent of inhibition of the receptor-kinase complex and not on the nature of a stimulus or on the type of the receptor involved.
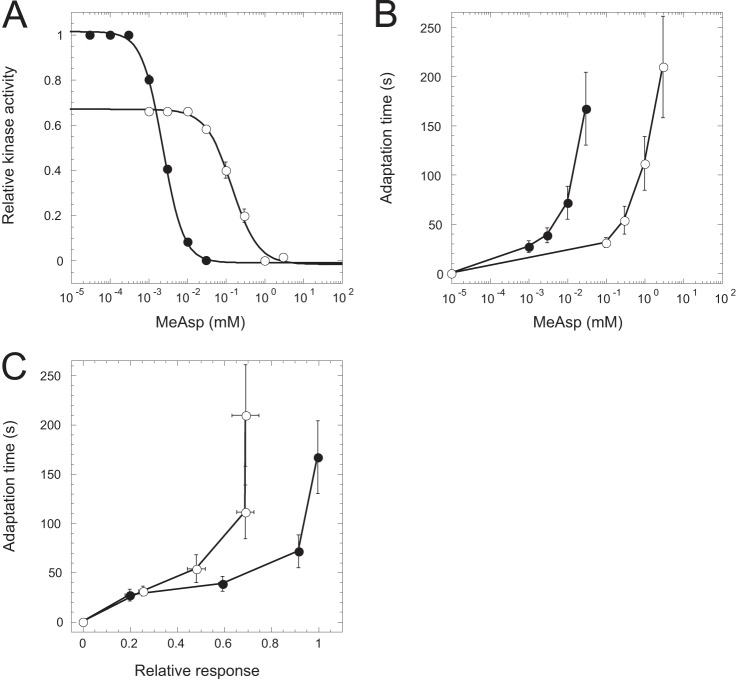
We further investigated whether the RA relation is affected by preadaptation to high background levels of attractant. Such preadaptation strongly increases receptor methylation (47, 48) and can slow the methylation kinetics due to partial saturation of methylation sites (43, 44). We therefore preadapted cells to 1 mM MeAsp, the ligand of the major receptor Tar, and measured the RA relation for stimulation starting from this ambient level. Consistent with previous reports (15, 17, 44), the dose dependence of the response amplitude (Fig. 2A) and that of the adaptation kinetics (Fig. 2B) were largely affected by preadaptation. Moreover, because the kinase activity does not fully recover upon adaptation to 1 mM MeAsp (44), the saturating response amplitude was smaller for the preadapted cells (Fig. 2A). For small response amplitudes, the RA alignment was nevertheless maintained even in the presence of high background stimulation (Fig. 2C).
FIG 2.
RA relation in wild-type cells in the presence of background attractant. Cells were stimulated with the indicated concentrations of MeAsp in the absence (filled circles) or presence (open circles) of 1 mM MeAsp in the background. The dependence of the relative kinase activity (A) and adaptation time (B) on the stimulus, as well as the RA relation (C), is shown.
RA relation in cells expressing individual receptors.
To better understand the nature of the observed universal alignment of the RA relation for different ligands, we investigated the responses of cells engineered to express only one type of receptor. We observed that when Tar or Tsr was expressed individually in the absence of other receptors, the RA relation strongly depended on the receptor type (Fig. 3A). Similar results were observed at induction levels that yield 2- to 3-fold differences in receptor expression (16, 49), confirming that the difference between the RA relations for Tar and Tsr is not due to their different expression levels. Compared to that in the wild type, the RA relation was particularly strongly affected in the Tsr-only cells. This is consistent with previous observations of inefficient adaptation to high levels of Tsr ligands (18, 44, 50) and of the serine-induced cross-methylation of Tar (51).
FIG 3.
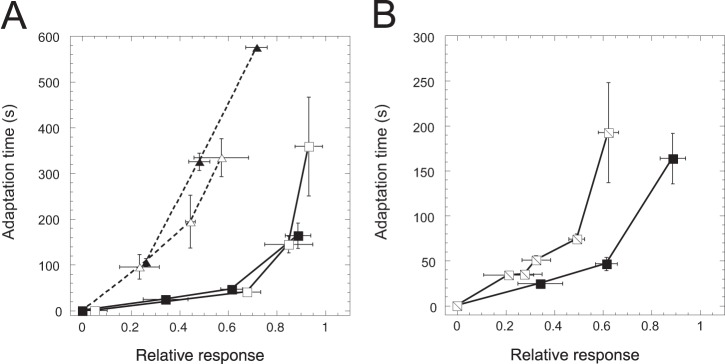
Dependence of RA relation in Tar-only or Tsr-only cells on receptor expression and on preadaptation. (A) RA relation for Tar-only cells induced with 1 μM Sal (open squares) or 2 μM Sal (filled squares) and in Tsr-only cells induced with 0.7 μM Sal (open triangles) or 1 μM Sal (filled triangles). (B) RA relation for Tar-only cells preadapted to buffer (filled squares, same as in panel A) or to 100 μM MeAsp (hatched squares).
Furthermore, the RA relation in strains expressing only one receptor showed a stronger dependence on the preadaptation to attractant (Fig. 3B). Although the level of prestimulation used for the Tar-only cells (100 μM MeAsp) is lower than the one used for the wild-type cells (1 mM) and is expected to lead to less methylation of Tar (compare Fig. S6A in reference 52 to Fig. S9 in reference 17), the adaptation kinetics in the preadapted Tar-only cells was consistently slowed down even for smaller stimuli. This is in contrast to the wild-type cells, where the slowdown occurred only at high levels of stimulation (Fig. 2C). Thus, the universal RA relation observed in wild-type cells is not an inherent property of the adaptation system.
Receptor interactions align the RA relation for different ligands.
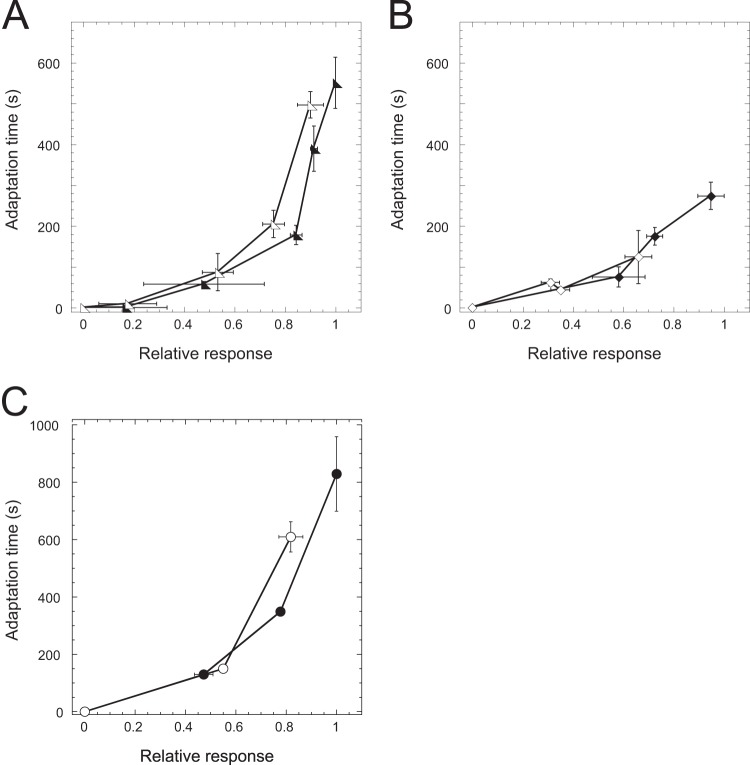
Instead, the alignment of the RA relation for different ligands can be easily explained by the Monod-Wyman-Changeux (MWC) model of allosteric interactions between different receptors in sensory complexes (12, 14, 16, 51, 53). Because these interactions couple the activity states of different receptors, the inactivation of one receptor by its ligand leads to the inactivation of other receptors in the same complex. Also, as an adaptation system acting on receptors depends on their activity, the resulting rate of adaptation should depend on the integral activity of the entire complex, irrespective of the type of the bound ligand. The allosteric coupling of receptor activities thus also effectively couples the rates of adaptation to different ligands. To test this prediction of the model, we first measured the RA relation in cells expressing either Tar or Tsr in the presence of other receptors. Indeed, under these conditions the RA relation for MeAsp and serine became realigned (Fig. 4A and B). The alignment was also observed when Tar was expressed in a strain that encoded only Tsr and lacked minor receptors (Fig. 4C), confirming that the coupling between Tar and Tsr is direct and does not require other receptors.
FIG 4.
Restoration of RA alignment by receptor coexpression. (A) RA relation for Δtar cells expressing Tar from a plasmid and stimulated with MeAsp (filled triangles) or serine (open triangles). (B) RA relation for tsr::Tn5 cells expressing Tsr from a plasmid and stimulated with MeAsp (filled diamonds) or serine (open diamonds). (C) RA relation for Δ(tar tap trg aer) cells expressing Tar from a plasmid and stimulated with MeAsp (filled circles) or serine (open circles). Receptor expression was induced with 1 μM Sal in all cases.
DISCUSSION
Two parameters of the bacterial chemotaxis pathway are central for the efficient navigation in chemoeffector gradients. One is the adapted value of the pathway output, CheY-P, which must fall into the narrow sensitive range of the flagellar motor (29, 32, 46, 54). Another parameter is the value of the adaptation rate, i.e., the rate of recovery of the kinase activity through changes in receptor methylation after the rapid initial response. The specific value of the adaptation rate is important for the chemotaxis strategy, because the time scale of the short-term memory needs to match the average time of runs performed by the bacteria navigating in a gradient (31–34). Faster adaptation would reduce the pathway stimulation in that the bacteria would only cover a short distance along the gradient before the memory is reset. Slower adaptation would lead to overstimulation of the pathway and ultimately disable temporal comparisons; in this case, the swimming direction would change during the measurement due to the rotational diffusion of the cell body. Such optimal adaptation rate should not depend on the type of the ligand, assuming that environmental gradients of these different ligand molecules have similar steepness.
The universal RA relation observed for all tested ligands is consistent with the existence of such an optimal adaptation rate. Here, we explored the RA relation only in the range of relatively strong stimuli and correspondingly long adaptation times. Nevertheless, the alignment of adaptation kinetics for strong stimulation is likely to apply equally to weak stimuli that are experienced by cells swimming in gradients, although the detailed shape of the RA relation might depend on stimulus strength (55). In principle, the RA alignment could be achieved through evolutionary fine-tuning of the receptor-specific rates of methylation and of the methylation effects on the receptor activity. However, we show that this is not the case and that individual receptor types exhibit very different RA relations. Instead, our results confirm theoretical predictions that the RA alignment for different ligands is ensured by the allosteric interactions of receptors within the sensory clusters. Such interactions are predicted to couple not only the activities of different receptors but also their methylation kinetics (51, 53), because the binding of an attractant molecule to any of the receptors in the allosteric signaling team reduces the activity of the entire team and therefore initially stimulates increased methylation of all receptors. Although due to the incomplete coupling between receptors such cross-methylation is only transient (51, 56), it is apparently sufficient to align the adaptation rates for different ligands in the case of the subsaturating stimulation in our experiments. Because the stimulation experienced by cells swimming in a gradient is even weaker than the stimuli used in our experiments, the observed RA alignment is highly relevant for chemotaxis.
Receptor interactions also ensure that the universal RA relation is maintained, at least for weak stimuli, upon adaptation to high background concentrations of attractants. This is consistent with a previous study (35) and is important because high levels of receptor methylation can decrease the addition rate of further methyl groups (43). The corresponding general decrease in the adaptation rate is indeed observed in the strain expressing only Tar. In wild-type cells, however, the allosteric coupling of Tar with Tsr and other receptors apparently provides additional free methylation sites, which can compensate for the lack of such sites on Tar and thus help to maintain the adaptation rate. Importantly, the observed mechanism of the RA alignment through allosteric coupling does not require any fine-tuning of the adaptation rates for individual receptors, which can change because of the growth stage dependence of receptor expression (17, 57, 58) or because of changes in the methylation levels of individual receptors.
The allosteric coupling of adaptation rates within receptor clusters is complementary to the previously proposed concept of adaptational assistance neighborhoods (24, 59, 60). The latter relies on binding of CheR and CheB to the C-terminal sequence of the major receptors, which allows these enzymes to methylate several receptors clustered in the vicinity of this tethering site. Such assistance is particularly important for proper modification of minor receptors that themselves do not possess the tethering sequence (61), and it may also play a role in enhancing the precision of adaptation for allosterically coupled receptors (62), but it does not by itself couple the adaptation kinetics of individual receptors. The proposed allosteric coupling thus represents yet another physiological function of receptor clustering in bacterial chemotaxis.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grant 294761-MicRobE from the European Research Council and by grant SO 421/11-1 from the Deutsche Forschungsgemeinschaft.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.02171-14.
REFERENCES
- 1.Levit MN, Liu Y, Stock JB. 1998. Stimulus response coupling in bacterial chemotaxis: receptor dimers in signalling arrays. Mol Microbiol 30:459–466. doi: 10.1046/j.1365-2958.1998.01066.x. [DOI] [PubMed] [Google Scholar]
- 2.Adler J. 1969. Chemoreceptors in bacteria. Science 166:1588–1597. doi: 10.1126/science.166.3913.1588. [DOI] [PubMed] [Google Scholar]
- 3.Hazelbauer GL, Falke JJ, Parkinson JS. 2008. Bacterial chemoreceptors: high-performance signaling in networked arrays. Trends Biochem Sci 33:9–19. doi: 10.1016/j.tibs.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hazelbauer GL, Lai WC. 2010. Bacterial chemoreceptors: providing enhanced features to two-component signaling. Curr Opin Microbiol 13:124–132. doi: 10.1016/j.mib.2009.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weis RM. 2006. Inch by inch, row by row. Nat Struct Mol Biol 13:382–384. doi: 10.1038/nsmb0506-382. [DOI] [PubMed] [Google Scholar]
- 6.Li M, Hazelbauer GL. 2011. Core unit of chemotaxis signaling complexes. Proc Natl Acad Sci U S A 108:9390–9395. doi: 10.1073/pnas.1104824108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ames P, Studdert CA, Reiser RH, Parkinson JS. 2002. Collaborative signaling by mixed chemoreceptor teams in Escherichia coli. Proc Natl Acad Sci U S A 99:7060–7065. doi: 10.1073/pnas.092071899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sourjik V. 2004. Receptor clustering and signal processing in E. coli chemotaxis. Trends Microbiol 12:569–576. doi: 10.1016/j.tim.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 9.Sourjik V, Armitage JP. 2010. Spatial organization in bacterial chemotaxis. EMBO J 29:2724–2733. doi: 10.1038/emboj.2010.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duke TAJ, Bray D. 1999. Heightened sensitivity of a lattice of membrane receptors. Proc Natl Acad Sci U S A 96:10104–10108. doi: 10.1073/pnas.96.18.10104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gestwicki JE, Kiessling LL. 2002. Inter-receptor communication through arrays of bacterial chemoreceptors. Nature 415:81–84. doi: 10.1038/415081a. [DOI] [PubMed] [Google Scholar]
- 12.Keymer JE, Endres RG, Skoge M, Meir Y, Wingreen NS. 2006. Chemosensing in Escherichia coli: two regimes of two-state receptors. Proc Natl Acad Sci U S A 103:1786–1791. doi: 10.1073/pnas.0507438103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li G, Weis RM. 2000. Covalent modification regulates ligand binding to receptor complexes in the chemosensory system of Escherichia coli. Cell 100:357–365. doi: 10.1016/S0092-8674(00)80671-6. [DOI] [PubMed] [Google Scholar]
- 14.Mello BA, Tu Y. 2005. An allosteric model for heterogeneous receptor complexes: understanding bacterial chemotaxis responses to multiple stimuli. Proc Natl Acad Sci U S A 102:17354–17359. doi: 10.1073/pnas.0506961102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sourjik V, Berg HC. 2002. Receptor sensitivity in bacterial chemotaxis. Proc Natl Acad Sci U S A 99:123–127. doi: 10.1073/pnas.011589998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sourjik V, Berg HC. 2004. Functional interactions between receptors in bacterial chemotaxis. Nature 428:437–441. doi: 10.1038/nature02406. [DOI] [PubMed] [Google Scholar]
- 17.Neumann S, Hansen CH, Wingreen NS, Sourjik V. 2010. Differences in signalling by directly and indirectly binding ligands in bacterial chemotaxis. EMBO J 29:3484–3495. doi: 10.1038/emboj.2010.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berg HC, Brown DA. 1972. Chemotaxis in Escherichia coli analysed by three-dimensional tracking. Nature 239:500–504. doi: 10.1038/239500a0. [DOI] [PubMed] [Google Scholar]
- 19.Macnab RM, Koshland DE Jr. 1972. The gradient-sensing mechanism in bacterial chemotaxis. Proc Natl Acad Sci U S A 69:2509–2512. doi: 10.1073/pnas.69.9.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Amin DN, Hazelbauer GL. 2010. The chemoreceptor dimer is the unit of conformational coupling and transmembrane signaling. J Bacteriol 192:1193–1200. doi: 10.1128/JB.01391-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goy MF, Springer MS, Adler J. 1977. Sensory transduction in Escherichia coli: role of a protein methylation reaction in sensory adaptation. Proc Natl Acad Sci U S A 74:4964–4968. doi: 10.1073/pnas.74.11.4964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boyd A, Simon MI. 1980. Multiple electrophoretic forms of methyl-accepting chemotaxis proteins generated by stimulus-elicited methylation in Escherichia coli. J Bacteriol 143:809–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Terwilliger TC, Wang JY, Koshland DE Jr. 1986. Kinetics of receptor modification. The multiply methylated aspartate receptors involved in bacterial chemotaxis. J Biol Chem 261:10814–10820. [PubMed] [Google Scholar]
- 24.Antommattei FM, Munzner JB, Weis RM. 2004. Ligand-specific activation of Escherichia coli chemoreceptor transmethylation. J Bacteriol 186:7556–7563. doi: 10.1128/JB.186.22.7556-7563.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perez E, West AH, Stock AM, Djordjevic S. 2004. Discrimination between different methylation states of chemotaxis receptor Tar by receptor methyltransferase CheR. Biochemistry 43:953–961. doi: 10.1021/bi035455q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barkai N, Leibler S. 1997. Robustness in simple biochemical networks. Nature 387:913–917. doi: 10.1038/43199. [DOI] [PubMed] [Google Scholar]
- 27.Banno S, Shiomi D, Homma M, Kawagishi I. 2004. Targeting of the chemotaxis methylesterase/deamidase CheB to the polar receptor-kinase cluster in an Escherichia coli cell. Mol Microbiol 53:1051–1063. doi: 10.1111/j.1365-2958.2004.04176.x. [DOI] [PubMed] [Google Scholar]
- 28.Kehry MR, Dahlquist FW. 1982. Adaptation in bacterial chemotaxis: CheB-dependent modification permits additional methylations of sensory transducer proteins. Cell 29:761–772. doi: 10.1016/0092-8674(82)90438-X. [DOI] [PubMed] [Google Scholar]
- 29.Lovdok L, Bentele K, Vladimirov N, Muller A, Pop FS, Lebiedz D, Kollmann M, Sourjik V. 2009. Role of translational coupling in robustness of bacterial chemotaxis pathway. PLoS Biol 7:e1000171. doi: 10.1371/journal.pbio.1000171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sourjik V, Wingreen NS. 2011. Responding to chemical gradients: bacterial chemotaxis. Curr Opin Cell Biol 24:262–268. doi: 10.1016/j.ceb.2011.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Andrews BW, Yi TM, Iglesias PA. 2006. Optimal noise filtering in the chemotactic response of Escherichia coli. PLoS Comput Biol 2:e154. doi: 10.1371/journal.pcbi.0020154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vladimirov N, Lovdok L, Lebiedz D, Sourjik V. 2008. Dependence of bacterial chemotaxis on gradient shape and adaptation rate. PLoS Comput Biol 4:e1000242. doi: 10.1371/journal.pcbi.1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oleksiuk O, Jakovljevic V, Vladimirov N, Carvalho R, Paster E, Ryu WS, Meir Y, Wingreen NS, Kollmann M, Sourjik V. 2011. Thermal robustness of signaling in bacterial chemotaxis. Cell 145:312–321. doi: 10.1016/j.cell.2011.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Celani A, Vergassola M. 2010. Bacterial strategies for chemotaxis response. Proc Natl Acad Sci U S A 107:1391–1396. doi: 10.1073/pnas.0909673107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lazova MD, Ahmed T, Bellomo D, Stocker R, Shimizu TS. 2011. Response rescaling in bacterial chemotaxis. Proc Natl Acad Sci U S A 108:13870–13875. doi: 10.1073/pnas.1108608108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shoval O, Goentoro L, Hart Y, Mayo A, Sontag E, Alon U. 2010. Fold-change detection and scalar symmetry of sensory input fields. Proc Natl Acad Sci U S A 107:15995–16000. doi: 10.1073/pnas.1002352107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Neumann S, Lovdok L, Bentele K, Meisig J, Ullner E, Paldy FS, Sourjik V, Kollmann M. 2014. Exponential signaling gain at the receptor level enhances signal-to-noise ratio in bacterial chemotaxis. PLoS One 9:e87815. doi: 10.1371/journal.pone.0087815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Steuer R, Waldherr S, Sourjik V, Kollmann M. 2011. Robust signal processing in living cells. PLoS Comput Biol 7:e1002218. doi: 10.1371/journal.pcbi.1002218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chalah A, Weis RM. 2005. Site-specific and synergistic stimulation of methylation on the bacterial chemotaxis receptor Tsr by serine and CheW. BMC Microbiol 5:12. doi: 10.1186/1471-2180-5-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Russo AF, Koshland DE Jr. 1983. Separation of signal transduction and adaptation functions of the aspartate receptor in bacterial sensing. Science 220:1016–1020. doi: 10.1126/science.6302843. [DOI] [PubMed] [Google Scholar]
- 41.Alon U, Surette MG, Barkai N, Leibler S. 1999. Robustness in bacterial chemotaxis. Nature 397:168–171. doi: 10.1038/16483. [DOI] [PubMed] [Google Scholar]
- 42.Berg HC, Tedesco PM. 1975. Transient response to chemotactic stimuli in Escherichia coli. Proc Natl Acad Sci U S A 72:3235–3239. doi: 10.1073/pnas.72.8.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meir Y, Jakovljevic V, Oleksiuk O, Sourjik V, Wingreen NS. 2010. Precision and kinetics of adaptation in bacterial chemotaxis. Biophys J 99:2766–2774. doi: 10.1016/j.bpj.2010.08.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Neumann S, Vladimirov N, Krembel AK, Wingreen NS, Sourjik V. 2014. Imprecision of adaptation in Escherichia coli chemotaxis. PLoS One 9:e84904. doi: 10.1371/journal.pone.0084904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sourjik V, Vaknin A, Shimizu TS, Berg HC. 2007. In vivo measurement by FRET of pathway activity in bacterial chemotaxis. Methods Enzymol 423:365–391. doi: 10.1016/S0076-6879(07)23017-4. [DOI] [PubMed] [Google Scholar]
- 46.Sourjik V, Berg HC. 2002. Binding of the Escherichia coli response regulator CheY to its target measured in vivo by fluorescence resonance energy transfer. Proc Natl Acad Sci U S A 99:12669–12674. doi: 10.1073/pnas.192463199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Springer MS, Goy MF, Adler J. 1979. Protein methylation in behavioural control mechanisms and in signal transduction. Nature 280:279–284. doi: 10.1038/280279a0. [DOI] [PubMed] [Google Scholar]
- 48.Sherris D, Parkinson JS. 1981. Posttranslational processing of methyl-accepting chemotaxis proteins in Escherichia coli. Proc Natl Acad Sci U S A 78:6051–6055. doi: 10.1073/pnas.78.10.6051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Endres RG, Oleksiuk O, Hansen CH, Meir Y, Sourjik V, Wingreen NS. 2008. Variable sizes of Escherichia coli chemoreceptor signaling teams. Mol Syst Biol 4:211. doi: 10.1038/msb.2008.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Masson JB, Voisinne G, Wong-Ng J, Celani A, Vergassola M. 2012. Noninvasive inference of the molecular chemotactic response using bacterial trajectories. Proc Natl Acad Sci U S A 109:1802–1807. doi: 10.1073/pnas.1116772109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lan G, Schulmeister S, Sourjik V, Tu Y. 2011. Adapt locally and act globally: strategy to maintain high chemoreceptor sensitivity in complex environments. Mol Syst Biol 7:475. doi: 10.1038/msb.2011.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Neumann S, Grosse K, Sourjik V. 2012. Chemotactic signaling via carbohydrate phosphotransferase systems in Escherichia coli. Proc Natl Acad Sci U S A 109:12159–12164. doi: 10.1073/pnas.1205307109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hansen CH, Sourjik V, Wingreen NS. 2010. A dynamic-signaling-team model for chemotaxis receptors in Escherichia coli. Proc Natl Acad Sci U S A 107:17170–17175. doi: 10.1073/pnas.1005017107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cluzel P, Surette M, Leibler S. 2000. An ultrasensitive bacterial motor revealed by monitoring signaling proteins in single cells. Science 287:1652–1655. doi: 10.1126/science.287.5458.1652. [DOI] [PubMed] [Google Scholar]
- 55.Shimizu TS, Tu Y, Berg HC. 2010. A modular gradient-sensing network for chemotaxis in Escherichia coli revealed by responses to time-varying stimuli. Mol Syst Biol 6:382. doi: 10.1038/msb.2010.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sanders DA, Koshland DE Jr. 1988. Receptor interactions through phosphorylation and methylation pathways in bacterial chemotaxis. Proc Natl Acad Sci U S A 85:8425–8429. doi: 10.1073/pnas.85.22.8425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kalinin Y, Neumann S, Sourjik V, Wu M. 2010. Responses of Escherichia coli bacteria to two opposing chemoattractant gradients depend on the chemoreceptor ratio. J Bacteriol 192:1796–1800. doi: 10.1128/JB.01507-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Salman H, Libchaber A. 2007. A concentration-dependent switch in the bacterial response to temperature. Nat Cell Biol 9:1098–1100. doi: 10.1038/ncb1632. [DOI] [PubMed] [Google Scholar]
- 59.Li M, Hazelbauer GL. 2005. Adaptational assistance in clusters of bacterial chemoreceptors. Mol Microbiol 56:1617–1626. doi: 10.1111/j.1365-2958.2005.04641.x. [DOI] [PubMed] [Google Scholar]
- 60.Li M, Hazelbauer GL. 2006. The carboxyl-terminal linker is important for chemoreceptor function. Mol Microbiol 60:469–479. doi: 10.1111/j.1365-2958.2006.05108.x. [DOI] [PubMed] [Google Scholar]
- 61.Barnakov AN, Barnakova LA, Hazelbauer GL. 1998. Comparison in vitro of a high- and a low-abundance chemoreceptor of Escherichia coli: similar kinase activation but different methyl-accepting activities. J Bacteriol 180:6713–6718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Endres RG, Wingreen NS. 2006. Precise adaptation in bacterial chemotaxis through “assistance neighborhoods.” Proc Natl Acad Sci U S A 103:13040–13044. doi: 10.1073/pnas.0603101103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.