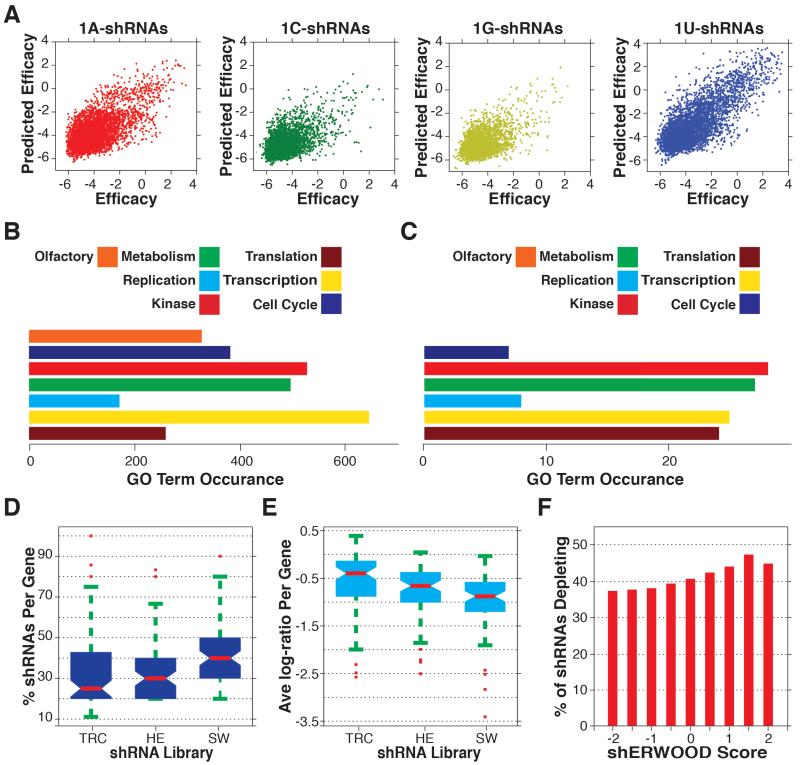
Figure 2. Construction and Validation of an shRNA-specific Predictive Algorithm.
A) Consolidated cross validation of predictions vs. sensor-scores for all shRNAs in the Fellmann et al. dataset (shRNAs are separated by the guide 5′ nucleotide). B) GO-term instances associated with the targeted gene set selected for shRNA validation screens. C) GO-term instances associated with genes for which at least two hairpins significantly depleted in each of the TRC, Hannon-Elledge (HE) and shERWOOD (SW) validation screens D) The percentage of shRNAs targeting consensus essential genes that depleted in each of the TRC, HE and shERWOOD shRNA screens. E) Average log-fold change for shRNAs targeting consensus essential genes (per gene) for each of the TRC, EH and shERWOOD validation screens. F) The percentage of shRNAs corresponding to consensus essential genes that, for any given shERWOOD score, depleted in the shERWOOD validation screen.