SUMMARY
Widespread quorum-sensing (QS) enables bacteria to communicate and plays a critical role in controlling bacterial virulence. However, effects of promiscuous QS crosstalk and its implications for gene regulation and cell decision-making remain largely unknown. Here we systematically studied the crosstalk between LuxR/I and LasR/I systems and found that QS crosstalk can be dissected into signal crosstalk and promoter crosstalk. Further investigations using synthetic positive feedback circuits revealed that signal crosstalk significantly decreases circuit’s bistable potential while maintaining unimodality. Promoter crosstalk, however, reproducibly generates complex trimodal responses resulting from noise-induced state transitions and host-circuit interactions. A mathematical model that integrates the circuit’s nonlinearity, stochasticity, and host-circuit interactions was developed, and its predictions of conditions for trimodality were verified experimentally. Combining synthetic biology and mathematical modeling, this work sheds light on the complex behaviors emerging from QS crosstalk, which could be exploited for therapeutics and biotechnology.
INTRODUCTION
Quorum-sensing (QS) is a widespread mechanism bacteria use to regulate gene expression and coordinate population behavior based on local cell density (Ng and Bassler, 2009). It is achieved through the binding of QS regulators with their cognate signal molecules (autoinducers) to regulate downstream QS pathways. Autoinducers are produced inside the cell and diffuse into and out of bacterial cells. Therefore, an autoinducer’s intracellular concentration correlates with local cell density (Ng and Bassler, 2009). There are diverse QS mechanisms allowing for bacterial communication: gram-positive bacteria generally use two-component systems mediated by peptides, while gram-negative bacteria primarily use LuxR/LuxI-type systems mediated by acylated homoserine lactones (AHL) (Miller and Bassler, 2001; Ng and Bassler, 2009). Many bacterial activities are controlled or regulated by QS, such as antibiotic production, biofilm development, bioluminescence, colonization, sporulation, symbiosis, and virulence (Jayaraman and Wood, 2008; LaSarre and Federle, 2013; Miller and Bassler, 2001; Ng and Bassler, 2009; Solano et al., 2014).
With well-defined and characterized biological properties, several QS regulators and corresponding autoinducers have also been used for synthetic gene networks. For example, LuxR/LuxI and/or LasR/LasI pairs were used to generate programmed patterns (Basu et al., 2005; Payne et al., 2013), trigger biofilm formation (Hong et al., 2012; Kobayashi et al., 2004), develop synthetic ecosystems and program population dynamics (Balagadde et al., 2008; Brenner et al., 2007), and construct synchronized oscillators (Danino et al., 2010; Prindle et al., 2012), edge detectors (Tabor et al., 2009), and pulse generators (Basu et al., 2004). RhlR/RhlI has also been used in the study of generic mechanisms of natural selection (Chuang et al., 2009) as well as for carrying out biological computations as chemical ‘wires’ (Tamsir et al., 2011).
However, effects of QS crosstalk, functional interactions between QS components that are not naturally paired, remain unexplored. For example, widely used LuxR-family regulators share extensive homologies and structural similarities in their corresponding autoinducers. LuxR binds its natural ligand 3-oxo-C6-HSL (3OC6HSL, hereafter denoted as C6) to activate the pLux promoter, while LasR bind 3-oxo-C12-HSL (3OC12HSL, hereafter denoted as C12) to activate pLas (Table S1) (Fuqua et al., 1996; Meighen, 1994; Miller and Bassler, 2001; Ng and Bassler, 2009; Schuster et al., 2004; Stevens and Greenberg, 1997). However, the LuxR protein can also bind other HSLs, such as C7HSL and 3OC8HSL (Canton et al., 2008). When binding C12, LasR is able to activate pLux in addition to the naturally paired pLas promoter (Balagadde et al., 2008). Implications of such crosstalk on gene regulation and cell response remain largely unknown.
Here, we use rationally designed gene networks to probe crosstalk between the LuxR/I and LasR/I systems and investigate their elicited bistable behaviors from positive feedback topologies. By using a synthetic biology approach, all combinations of autoinducer, regulator, and promoter were tested to show that QS crosstalk can be dissected into signal crosstalk and promoter crosstalk. When studied in the context of a synthetic positive feedback gene network, our results indicate that QS crosstalk leads to distinct dynamic behaviors: signal crosstalk significantly decreases the circuit’s induction range for bistability, but promoter crosstalk causes transposon insertions into the regulator gene and yields trimodal responses due to a combination of mutagenesis and noise induced state transitions. To fully understand this complex response, we developed and experimentally verified a mathematical model that takes into account all of these factors to simulate and predict how varying the transposition rate can modulate this trimodality. This reveals a novel factor of host-circuit interactions in shaping complex responses of synthetic gene networks.
RESULTS
Dissecting the crosstalk between LuxR/I and LasR/I using synthetic circuits
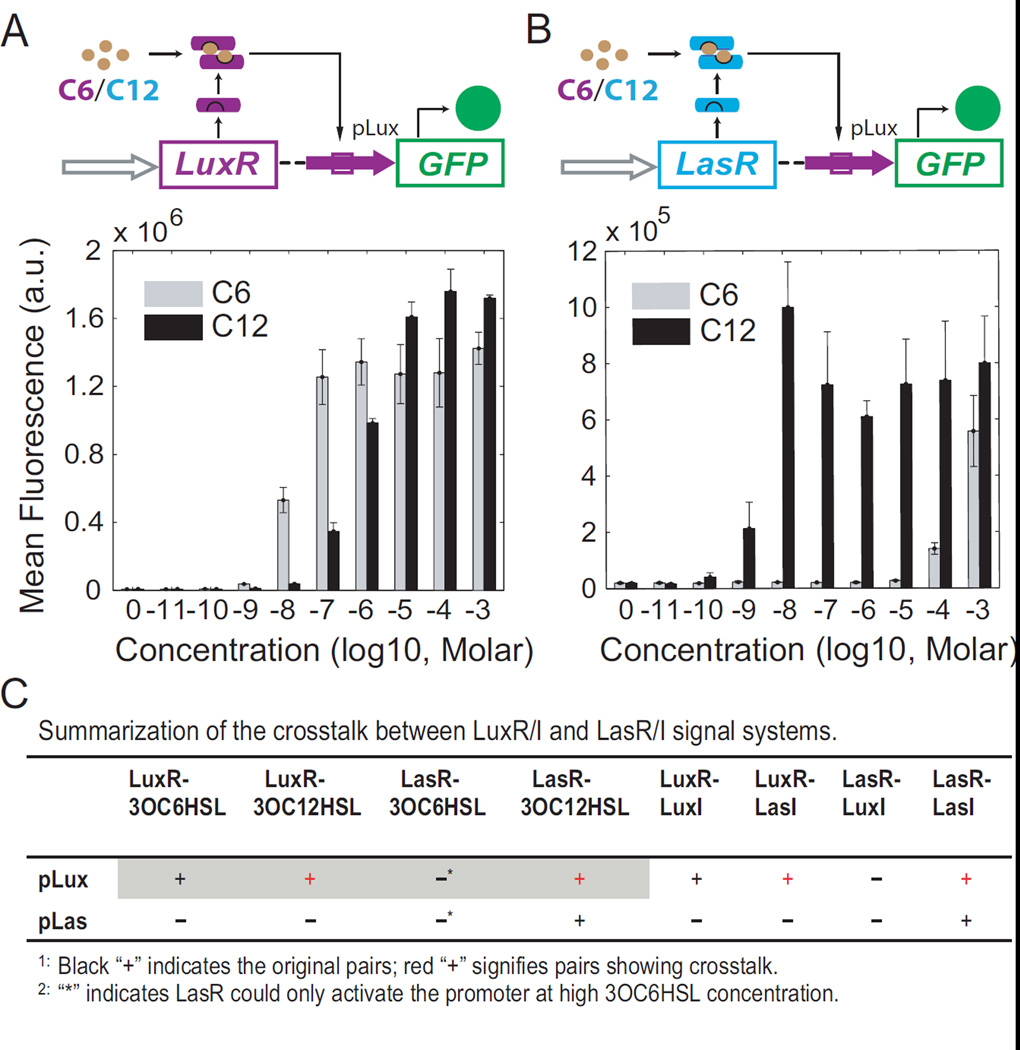
To characterize possible crosstalk between LuxR/I and LasR/I signaling systems, four synthetic circuits, CP (constitutive promoter)-LuxR-pLux (Figure 1A), CP-LasR-pLux (Figure 1B), CP-LasR-pLas (Figure S1A), and CP-LuxR-pLas (Figure S1B), were first built to test all autoinducer-regulator-promoter combinations’ impact on gene expression activation. C6 and C12 were applied independently to all constructs, and green fluorescent protein (GFP) expression under the regulation of pLux or pLas was measured as the readout.
Figure 1. QS crosstalk dissected using synthetic gene circuits.
(A) LuxR can crosstalk with C12 to activate pLux. Top panel: schematic diagram of a synthetic gene circuit where a constitutive promoter (gray arrow) regulates LuxR (purple rectangle) expression. LuxR protein (purple bars), when dimerized and bound with C6 or C12, can activate pLux (purple arrow) to induce GFP (green rectangle) expression. The autoinducers, genes, and promoters are color coded so that naturally paired partners are in the same color. Bottom panel: dose response of the circuit when induced with C6 (gray) or C12 (black). (B) LasR can crosstalk with pLux when bound with C12. Top panel: schematic diagram of a circuit similar to that in (A), where a constitutive promoter regulates LasR (cyan rectangle) expression. LasR protein, when bound with C6 or C12, can activate pLux to induce GFP expression. Bottom panel: Dose response of this circuit when induced with C6 (gray) or C12 (black). Bar heights are averages of three independent flow cytometry measurements shown as mean ± SD. (C) Summary of crosstalk induction of all 16 different combinations, including inductions by both chemicals and corresponding synthase genes. The four combinations shown in (A) and (B) are highlighted with a gray background. Quantified results for other combinations are included in Figure S1.
It can be seen in Figure 1A that in addition to its natural partner C6, LuxR can also bind 5 with C12 molecules to activate pLux, which suggests that the binding with C6 or C12 results in a similar conformational change of LuxR and therefore its activating functions remain uninterrupted. Such an activation of a natural QS regulator-promoter pair by a cross-talking autoinducer is here termed signal crosstalk. It can be seen that this signal crosstalk can fully activate the system with comparable induction dosages. However, similar tests of signal crosstalk of C6 with the Las regulator-promoter pair (Figure S1A) only show comparable induction when the autoinducer concentration is as high as 10−3 M. This suggests that the efficacy of signal crosstalk is QS system specific.
In addition to promiscuous autoinducer binding resulting in signal crosstalk, the systems studied also displayed crosstalk between regulators and promoters, here termed promoter crosstalk. It is shown in Figure 1B that, in addition to being able to activate pLas, LasR significantly activates pLux when induced with its natural cognate ligand C12, though not with C6, which suggests that LasR’s DNA binding domain can recognize both pLas and pLux when bound with its natural partner. This promoter crosstalk is robust over a wide range of autoinducer concentrations. Similar tests of promoter crosstalk of C6-LuxR to pLas (Figure S1B) show only weak induction. This suggests that the efficacy of promoter crosstalk is also QS system specific. It should also be noted that a third type of crosstalk, regulator crosstalk, in which naturally paired autoinducer and promoter function through a cross-talking regulator protein, only exhibited minimal levels of activation (gray bar in Figure 1B and black bar in Figure S1B).
To further verify the crosstalk under physiologically relevant dosages of autoinducers, synthase genes LuxI and LasI were introduced to replace commercial chemicals in eight different circuits (Figure S1C and 1D). The results further confirm that pLux can be activated by LuxR with LuxI or LasI, as well as by LasR with LasI. This is consistent with the results using commercial chemicals, indicating the crosstalk categorization is also applicable in vivo. All combinatorial activations between LuxR/I and LasR/I systems are summarized in Figure 1C, with crosstalk highlighted in red. Taken together, detectable crosstalk between LuxR/I and LasR/I systems can be categorized into two types: LasI (C12) can crosstalk with the LuxR protein to induce pLux transcription (signal crosstalk), and the LasR-LasI (C12) complex can also crosstalk with and activate the pLux promoter (promoter crosstalk).
Signal crosstalk induces distinct responses from positive feedback circuits
Next, synthetic positive feedback circuits were constructed to investigate the impact of QS crosstalk in the context of gene regulatory networks. It is shown that the core of many bacteria’s QS decision-making circuits is a positive feedback motif (Ji et al., 1995; Kaplan and Greenberg, 1985; de Kievit and Iglewski, 2000; Pestova et al., 1996; Piper et al., 1993; Seed et al., 1995). Because of its potential bistability, such a topology enables the bacteria to make appropriate binary decisions in response to changing environments (Ozbudak et al., 2004; Xiong and Ferrell, 2003; Guido et al., 2006; Isaacs et al., 2003). Synthetic positive feedback circuits serve as suitable platforms to probe the effects of signal and promoter crosstalk within the framework of gene regulatory networks.
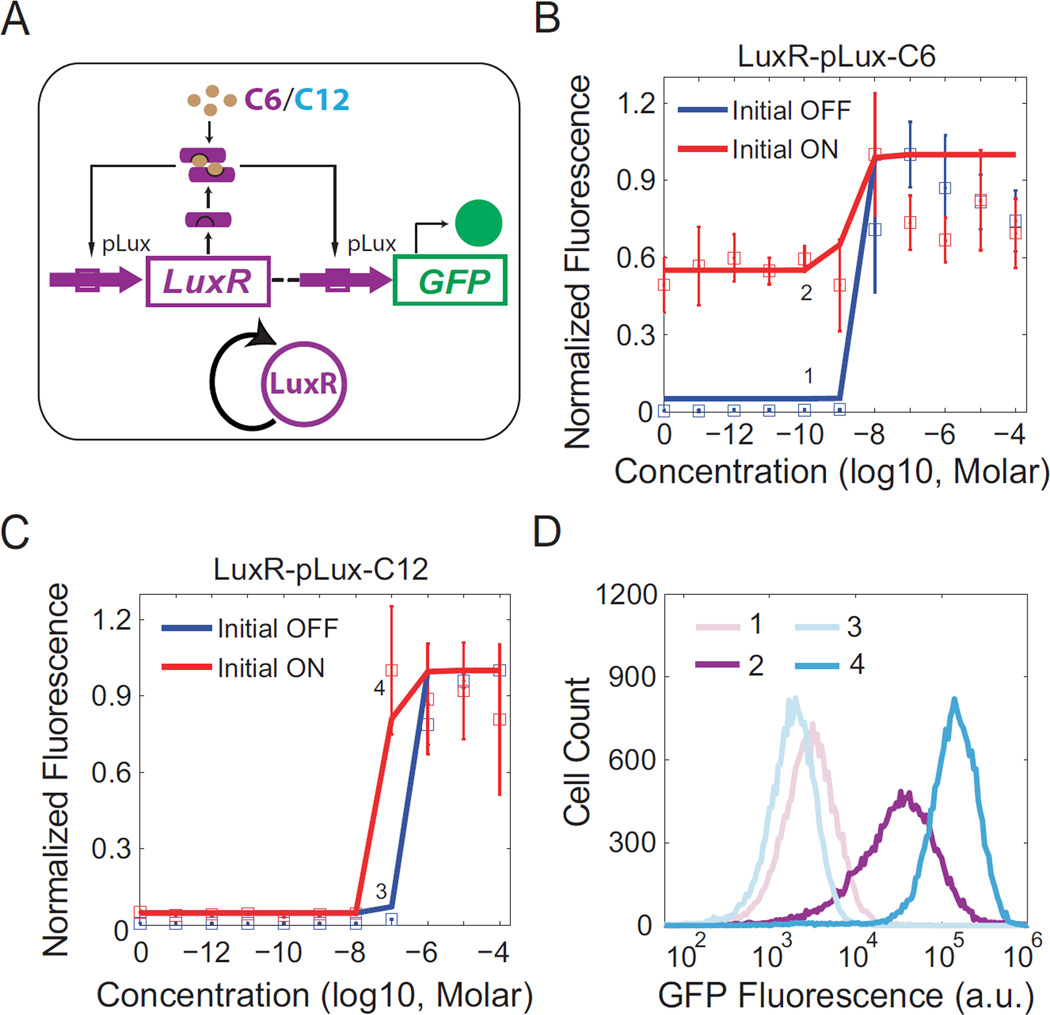
The design shown in Figure 2A was first constructed to study signal crosstalk. In this circuit, expression of LuxR is regulated by the promoter pLux, which can be activated by LuxR when induced, forming a positive feedback loop. pLux driven GFP expression serves as the readout for LuxR levels. Robustness of history-dependent responses (hysteresis), a hallmark of many positive feedback topologies, is used as the main measure of signal crosstalk impacts as it captures the effectiveness of the circuit’s decision-making functionality (Acar et al., 2005; Gardner et al., 2000; Wu et al., 2013).
Figure 2. Signal crosstalk causes shrinkage of bistable region.
(A) Schematic diagram of a synthetic gene circuit where the pLux promoter regulates expression of LuxR, which in turn can bind with C6 or C12 to further activate pLux, forming a positive feedback loop (shown as simplified diagram). GFP under the regulation of pLux serves as the readout for LuxR levels. All components are color coded similarly as in Figure 1. (B) The average of three replicate flow cytometry measurements is plotted as a square with error bars for each dose of C6 induction, where red indicates Initial ON cells while blue denotes Initial OFF cells. Solid lines represent results calculated from model fittings. The bistable region ranges from 0 to 10−9 M C6. Labels 1 and 2 indicate representative experiments within the region to be shown as histograms in (D). (C) Similar experiments as in (B) but with C12 inductions. The bistable region ranges from 10−8 to 10−6 M C12. Labels 3 and 4 indicate representative experiments within the bistable region to be shown as histograms in (D). (D) Histograms of flow cytometry measurements labeled in (B) and (C). One representative measurement from each point is shown. No bimodal distributions are observed.
As a benchmark, uninduced (Initial OFF) cells with the circuit were first induced with different concentrations of LuxR’s natural inducer C6 and measured using flow cytometry (Figure 2B, blue). It can be seen that GFP is only turned on with 10−8 M or higher C6 induction. The cells treated with 10−4 M C6 (Initial ON) were then collected and diluted into new medium with the same concentrations of C6 (Figure 2B, red). These cells keep high GFP expression even with low C6 inductions (below 10−9 M) due to the self-sustaining nature of positive feedback loops. Taken together, these results illustrate this circuit’s hysteretic response with C6 inducer concentrations between 0 and 10−8 M. This indicates that under C6 induction the positive feedback circuit is bistable between 0 and 10−8 M C6 induction. However, no bimodal distribution was observed within the bistable region based on flow cytometry measurements (Figure 2D, purple and light purple; and Figure S2A), suggesting that the barrier between the two states is too high for inherent gene expression stochasticity to overcome (Acar et al., 2005; Gardner et al., 2000).
Next, C12 was used to induce the same construct to investigate the impact of signal crosstalk on gene network regulation. Similar induction experiments were carried out and the results are shown in Figure 2C. It can be seen that this circuit also displays hysteresis, but with a much smaller bistable region between 10−8 and 10−6 M C12. Flow cytometry results within the bistable region also show no bimodal distributions (Figure 2D, cyan and light cyan; and Figure S2B).
To quantitatively understand the signal crosstalk caused shrinkage of the bistable region, an ordinary differential equation (ODE) model of LuxR-pLux auto-activation was developed. Two major kinetic events, LuxR transcription and translation, are described by two ODEs with all binding between chemical species incorporated into model terms. After fitting the parameters using existing literature and experimental measurements (Table S2), the model can capture the experimental results (lines in Figure 2B and 2C) with accuracy. Inspection of model parameters reveals that the bistable region decrease caused by signal crosstalk can be largely accounted for by differential binding affinities between LuxR and C6 and C12. This suggests a new way to perturb QS decision-making through utilization of crosstalking autoinducers, which could be useful for clinical therapies.
Promoter crosstalk induces unexpected and complex bimodal responses
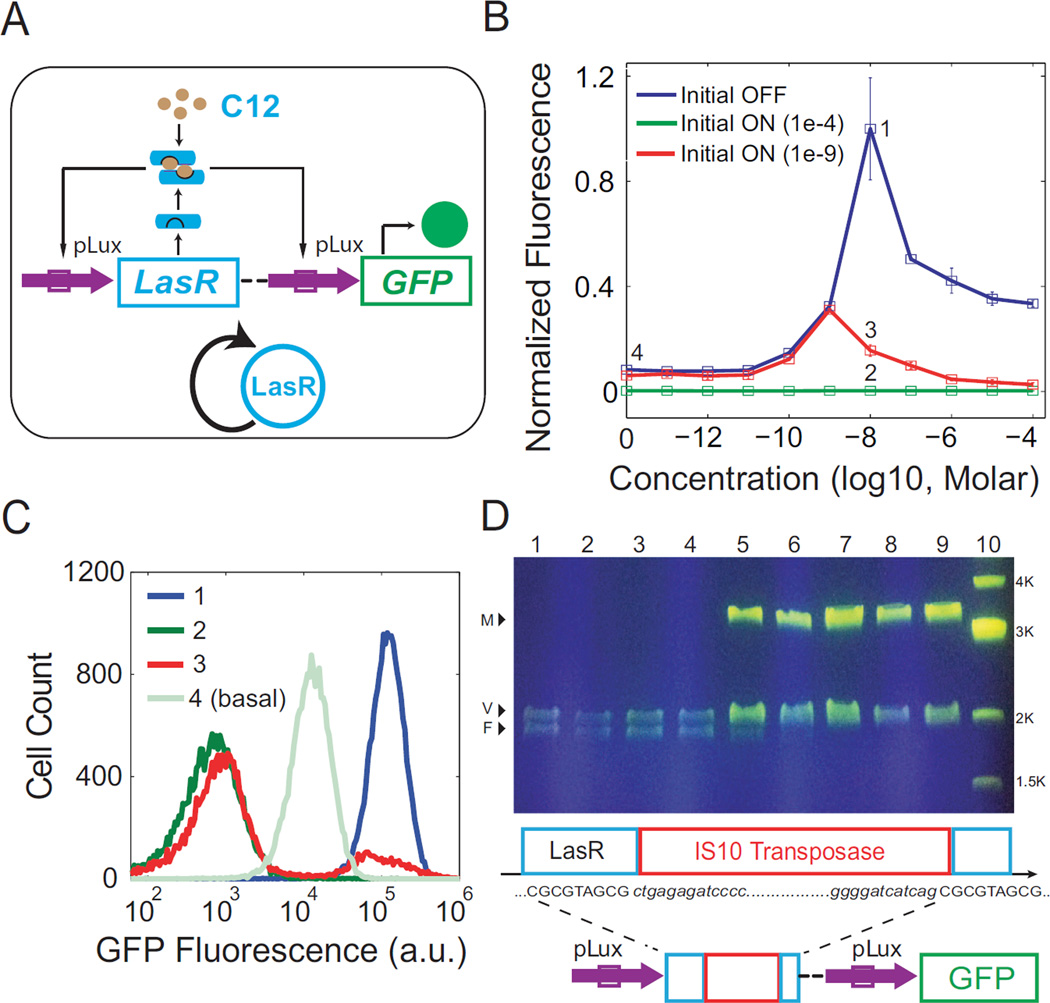
To study the impacts of promoter crosstalk, a positive feedback circuit was constructed with LasR under the regulation of pLux (Figure 3A). It is shown in Figure 1B that LasR can activate pLux when induced by C12. Therefore this circuit also forms a positive feedback loop in the presence of C12. GFP under regulation of pLux is again included as a readout for LasR. Experimental explorations of hysteresis were carried out and the results are shown in Figure 3B. It can be seen that initial OFF cells (blue) exhibit a non-monotonic response to C12 induction: GFP expression increases with C12 concentration, but begins to uniformly decrease when C12 induction exceeds 10−8 M (Figure 3B, and Figure S3A and 3B). Cells induced with 10−4 M C12 were then collected and diluted into fresh medium with the same inducer concentrations as the initial OFF cells. Flow cytometry data show that all samples exhibit unimodal minimal fluorescence signals that are even lower than the basal GFP expression of initial OFF cells (Figure 3B and 3C green, and Figure S3B).
Figure 3. Promoter crosstalk induces mutation and leads to population heterogeneity.
(A) Schematic diagram of a synthetic LasR-pLux positive feedback circuit. GFP under the regulation of pLux serves as the readout for LuxR levels. All components are color coded similarly to Figure 1. (B) The average of three replicate flow cytometry measurements is plotted as a square with error bars for each dose of C12 induction. Blue denotes Initial OFF cells, while green and red indicate the Initial ON cells induced with 10−4 M C12 and 10−9 M C12 before being re-diluted into concentrations of C12, respectively. Labels 1, 2, 3, and 4 indicate experiments to be shown in detail as histograms in (C). (C) Histograms of flow cytometry measurements labeled in (B). One representative measurement from each point is shown. A bimodal distribution is only observed for label 3: which is Initial ON cells (induced with 10−9 M C12 before redilution) at 10−8 M C12. (D) DNA analysis for the Initial ON samples shown as red in (B). Top: Plasmid DNA was extracted and digested with EcoRI and PstI, and argarose gel electrophoresis results indicated gene mutation happened in samples with 10−8 M and higher doses of C12. Lane 1 is the wild-type plasmid as the control, lanes 2 to 9 are samples in 10−11 to 10−4 M C12, and Lane 10 is the 1kb DNA marker. V: vector; F: wild-type DNA fragment (the LasR-pLux positive feedback circuit); M: mutated fragment. Bottom: Schematic representation of the mutation and the features of IS10 transposase insertion: the target site (first CGCGTAGCG) in the LasR gene, its duplication (second CGCGTAGCG) due to insertion of IS10 transposase, and the IS10 sequence (red box and shown in italics).
Considering that both C12 and exogenous gene overexpression may be toxic to cells, as well as the fact that initial OFF cells can be turned on with lower induction dosages, cells induced with lower than 10−4 M but higher than 10−10 M C12 were collected as new initial ON cells to further explore possible hysteresis of this circuit. Collected cells were diluted into fresh medium with the same concentrations of C12. These new initial ON cells demonstrate the same expression pattern as the initial OFF cells when grown in inducer concentrations from 0 to 10−9 M, but they show much lower fluorescence values at higher concentrations. For example, the red points in Figure 3B illustrate the GFP average of 10−9 M induced initial ON cells when collected and re-diluted into a range of C12 concentrations (See Figure S3C for results with other initial induction dosages). Examination of the flow cytometry measurements of these ON cells reveals that bimodal distributions emerge within the concentration range of 10−8 M to 10−4 M C12. Interestingly, one peak of the distribution is at the high state and the other is at the minimal expression state, even lower than basal expression (Figure 3C, red). So unlike classic bimodal responses due to bistability, LasR-pLux positive feedback exhibits bimodality with the lower peak’s expression even weaker than the OFF state. To exclude the possibility that this bimodality is triggered by inherent properties of the LasR-C12 complex, similar hysteresis experiments were carried out for the linear CP-LasR-pLux circuit (Figure 1B). Results show that the initial OFF and ON cells both exhibit unimodal expression without hysteresis (Figure S3D). The bimodality is, therefore, unique to the initial ON cells with LasR-pLux positive feedback.
Bimodality results from circuit-host interactions
The remaining question is: what is the cause of the minimal expression state? To resolve this problem, new initial ON samples at concentrations of 10−11 M to 10−4 M C12 (Figure 3B, red triangles) were collected. Their plasmids were extracted and digested for genotyping. The agarose gel electrophoresis results show that a new band (~3.2 kb) replaces the original fragment band (wild type, ~1.9 kb) for samples in 10−8 ~10−4 M C12, and that a faint original-fragment band can also be seen for samples with 10−8 and 10−7 M C12 inductions (Figure 3D). Further sequencing analyses verify that an IS10 transposase is inserted into the LasR gene at the 682 bp site and this insertion is flanked by two 9 bp direct repeats 5’-CGCGTAGCG-3’ (Figure 3D and Supplementary Information), which is consistent with reported hotspots for IS10 insertion (Kovarík et al., 2001).
The insertion abolishes LasR’s ability to activate downstream GFP expression, which in turn causes the cells’ fluorescence signal to be even weaker than basal expression when LasR is intact. Cells with this type of mutation form the low GFP peak in the bimodal distributions in Figure 3C. On the other hand, cells that do not mutate are able to maintain a high GFP expression due to positive feedback, forming the GFP ON peak of the bimodal distributions. Taken together, the combination of gene network activated GFP expression and mutation caused GFP inhibition drive the emergence of a bimodal distribution.
Trimodality predicted by expanded model
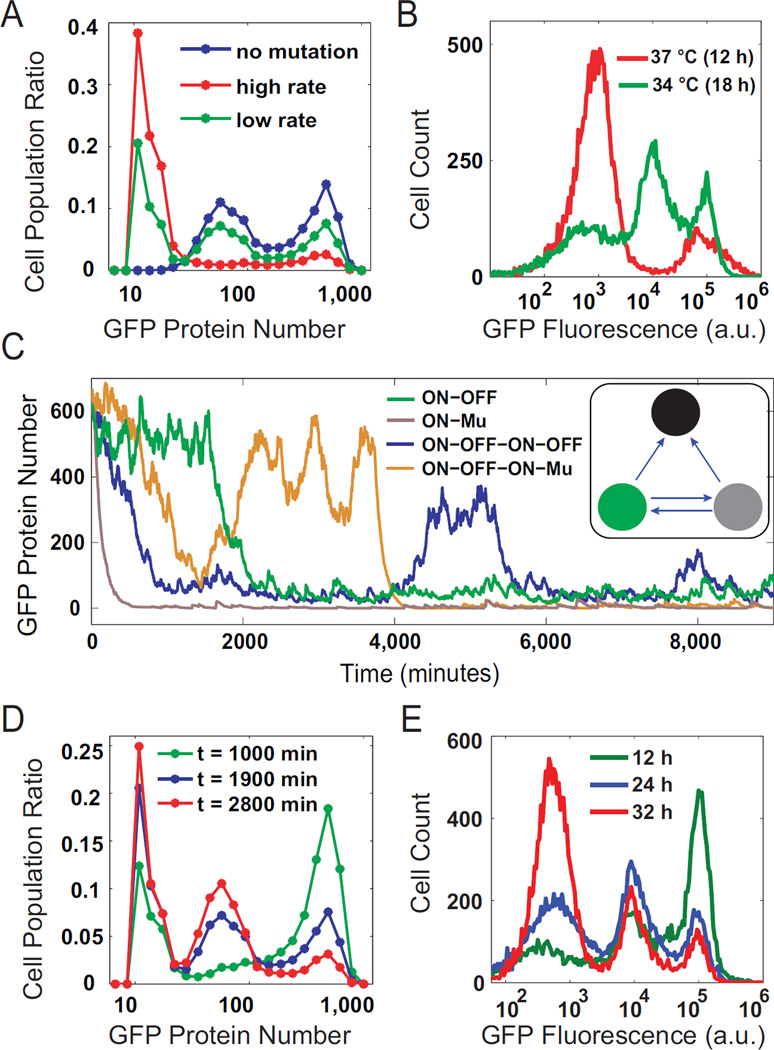
In light of the verified mutation in the LasR-pLux positive feedback system, the mathematical model was expanded to take into account crosstalk triggered genetic changes to better describe the circuit. To enable comparison with flow cytometry results, the ODEs were transformed into corresponding biochemical reactions and simulated stochastically (Gillespie, 1977). In addition, each cell was assigned a probability of mutation throughout the simulation (Figure 4C inset), which is dependent on the cell’s current LasR/GFP level and the transposition rate. Once mutated, the cells had only minimal GFP expression strength and remained mutated until the end of the simulation. Finally, growth rate differences between wild type and mutated cells were computed from experiments (Figure S4A) and taken into consideration in the simulation. Results of stochastic simulations of this expanded model are shown in Figure 4A, exhibiting the bimodal distribution observed experimentally (red curves in Figure. 4A, simulation; and 4B, experiment).
Figure 4. Model predictions and experimental validations of mutation induced trimodality.
(A) Model predictions of GFP expression at several transposition rates: high (red, k3=3.6e-6), low (green, k3=4e-7), and none (blue, k3=0). Histograms were constructed from 8000 single cell stochastic simulations at 1000 (k3=3.6e-6) and 1900 (k3=0 and k3=4e-7) minutes. (B) Experimental validation of the model predictions in (A). Red and green curves correspond to the high and low transposition rates from (A), and they exhibit similar bi- and trimodal responses, respectively. No blue curve is included because mutation could not be eliminated entirely experimentally. (C) Representative stochastic simulations of single cell fluorescence starting from the ON state. All possible transitions are shown. Inset diagram illustrates all possible state transitions in the simulation. (D) Model predictions of GFP expression with low transposition rate showing temporal evolution of the population from primarily ON cells at an early time (green), to trimodal distributions at intermediate time (blue), eventually falling into a primarily Mutated state at late time (red). (E) Flow cytometry measurements taken at 12 hours (green), 24 hours (blue), and 32 hours (green). Populations show similar dynamics to those predicted by the model in (D), starting with a large ON peak, transitioning to a trimodal distribution, then into primarily Mutated or OFF cells.
To further investigate the impact of this mutation on the circuit’s functions, simulations were carried out with perturbed parameters to mimic various scenarios. First, the transposition rate was artificially set to zero, and the simulations show that the system can also exhibit a bimodal distribution (Figure 4A, blue), with the OFF peak exhibiting basal GFP expression. Bimodality has been reported to arise from stochastic state switching of a bistable system without any genetic changes (Acar et al., 2005; Gardner et al., 2000; Tan et al., 2009). The same mechanism leads to simulated bimodality of this LasR-pLux circuit when there is no mutation.
While it is almost impossible to eliminate mutation, it is possible to decrease the transposition rate experimentally. To explore the impacts of mutation in a more realistic scenario, simulations were carried out with positive but smaller transposition rates. Interestingly, the system demonstrates a trimodal distribution (Figure 4A, green). In this distribution, there are three groups of cells: ON, OFF, and Mutated. Those cells initialized at the ON state freely transition to and from the OFF state, due to the system’s bistability. Meanwhile, all cells have the chance to mutate and stay mutated (Figure 4C). Given enough time and the right measurement window, all three groups of cells would be visible. Within this window, the portion of ON and OFF cells will gradually decrease and the number of mutated cells will increase because the mutation is irreversible. The effect of a decreased transposition rate is essentially slowing down the ON to Mutation transition rate and giving enough time for ON to OFF transitions and hence the emergence of the OFF peak. Time courses of the simulations demonstrate gradual emergence and evolution of these three populations of cells (Figure 4D).
Experimental validation of trimodal responses by lowering growth temperature
Previous reports indicated that transposition frequency can be perturbed by growth temperatures (McClintock, 1984; Ohtsubo et al., 2005; Sousa et al., 2013). To tune the transposition rate, experiments were carried out with cells cultured at a lower 34 °C temperature, which was shown to slow down crosstalk triggered mutation of this circuit (Figure S4B). Consistent with model predictions, initial ON cells induced with 10−8 M C12 exhibited a trimodal response when the growth temperature was tuned from 37 °C to 34 °C (Figure 4B, green). Moreover, temporal evolution of the proportion of each subpopulation was consistent with model predictions: the portion of ON cells gradually decreased, the Mutation portion increased, and the OFF portion increased first and then decreased as time went on (Figure 4E). Growth rates of cells at Mutated, ON, or OFF states were also measured and show no difference when cultured at these two different temperatures (Figure S4A). The emergence of the OFF peak, therefore, is fully accounted for by the decrease of transposition rate, which slows down the direct transitions from ON to Mutation and therefore gives the cells time to layover at the OFF state. This is also evidenced by the smaller portion of Mutated cells when grown at 34 °C compared with 37 °C (Figure S4B).
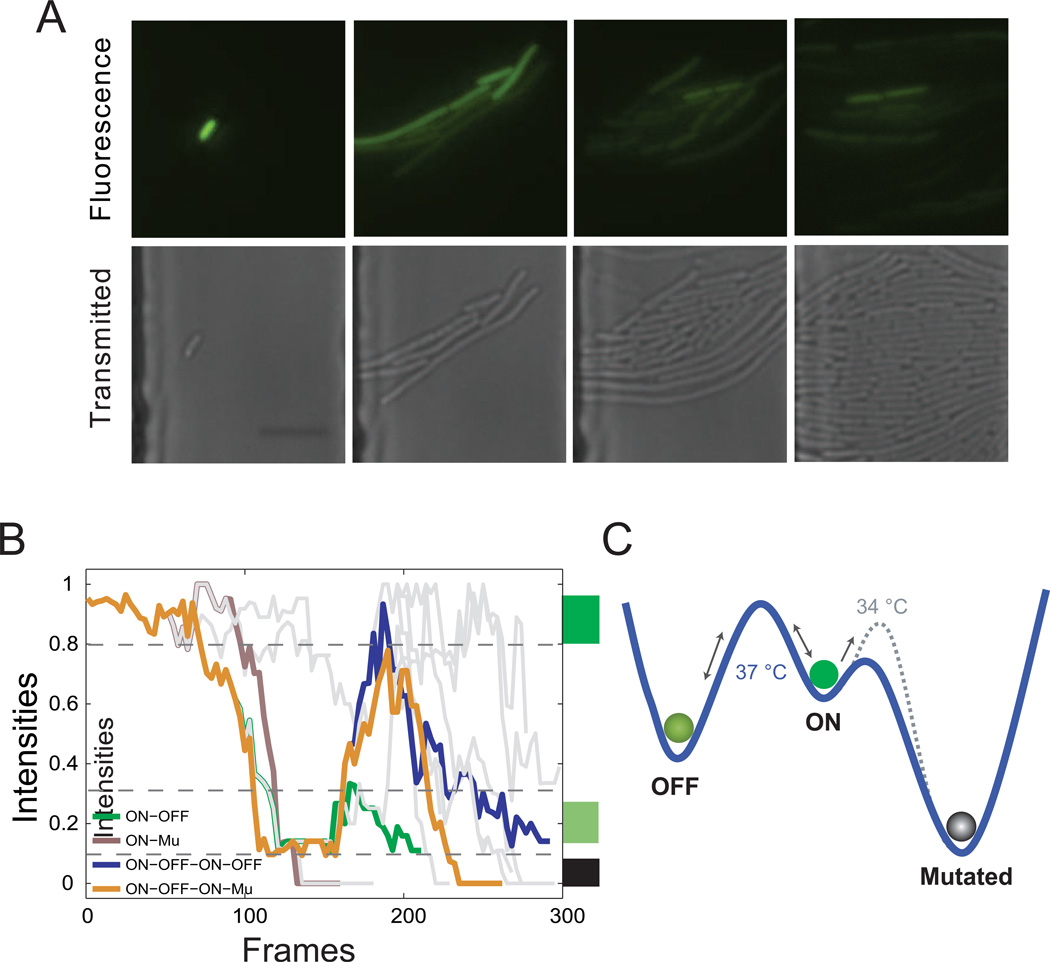
Furthermore, a microfluidic platform coupled with time-lapse imaging was also employed to verify model predictions (Ferry et al., 2011). Cells were pretreated with 10−9 M C12 until steady state as the initial ON cells before being loaded into the device and induced with 10−8 M C12 at 34 °C to mimic the experimental protocols used in Figure 4E. Initially, there was only one ON cell loaded into the trap (Figure 5A and Movie S1). At the 8th hour, it can be seen that two populations began to emerge: some cells became OFF and some stayed ON. Mutations started to occur shortly after the 8th hour, and the OFF and Mutation cells accounted for around 90 percent of the population after 16 hours. Eventually mutation state cells took up the majority of the population. There also existed several OFF cells which became ON again, owing to stochastic gene expression noise, but they eventually exhibit a similar evolving process: ON to OFF or Mutation (Figure 5B and Movie S1), which is consistent with the stochastic model simulations shown in Figure 4C.
Figure 5. Fluorescence microscopy validation of mathematical model predictions.
(A) GFP fluorescence (top) and phase contrast (bottom) images of cells growing in the microfluidic chamber at 0, 8, 16, and 24 hours. Magnification: 40×. (B) Normalized fluorescence expression of representative cells from (A), showing similar behavior to that predicted by the model from Figure 4C. Four cells are colored corresponding to the scenarios in Figure 4C, and the other 11 cells are grey. Each trajectory follows one cell, with the trajectory branching as the cells divide. One frame equals five minutes. (C) Diagram of the mechanism for trimodality. Each “valley” represents one state. The blue curve represents the landscape at 37 °C, and the dotted grey curve is the landscape at 34 °C. At 37 °C, ON state cells can more easily transition to the Mutated state because of the low barrier; while at 34 °C, the barrier between ON and Mutated states increases, resulting in more ON cells transitioning to OFF state and promoting the emergence of trimodality.
Altogether, the flow cytometry and microfluidic data confirmed the model’s predicted trimodality, which arises from bistability of the positive feedback circuit and host-circuit interactions. In the context of positive feedback, cells transition freely between the ON and OFF states, but it is easier for ON state cells to transition to the OFF state because of the asymmetric energy barrier (Figure S4C). However, the ON cells can also transition to the Mutated state, which carries an advantage of growth rate (Figure S4A). Compared to OFF state cells, those in the ON state would transition more frequently to the Mutated state at 37 °C, leading to the bimodal distribution (Figure 3). When the growth temperature is reduced to 34 °C, the transposition frequency also decreases, meaning that the barrier between ON and Mutated state increases. Therefore, more ON cells would transition to the OFF state, which promotes the emergence of trimodality (Figure 5C).
DISCUSSION
QS is a ubiquitous mechanism in nature, and its regulator-autoinducer pairs, such as LuxR/LuxI and LasR/LasI, have been used in synthetic biology for a wide range of applications (Balagadde et al., 2008; Basu et al., 2004, 2005; Brenner et al., 2007; Chen et al., 2014; Chuang et al., 2009; Danino et al., 2010; Hong et al., 2012; Kobayashi et al., 2004; Payne et al., 2013; Prindle et al., 2012; Tabor et al., 2009; Tamsir et al., 2011; Pai et al., 2012). However, evolutionary pressures from limited resources in a competitive environment promote promiscuous bacterial communication, which takes the form of either different genera of bacteria producing the same types of autoinducers or non-specific regulator-autoinducer binding (Balagadde et al., 2008; Gray et al., 1994; Hong et al., 2012; Miller and Bassler, 2001; Pérez et al., 2011; Winzer et al., 2000). As a result, QS regulator-autoinducer pairs are not orthogonal, and there is crosstalk between them. Dissecting the crosstalk is critical for unraveling the underlying principles of bacterial decision-making and survival strategies for both natural and synthetic systems.
In this work, we used synthetic biology approaches to dissect QS crosstalk between LuxR/I and LasR/I. By applying engineering principles to construct modular gene networks, we were able to characterize and categorize QS crosstalk into signal crosstalk, where LuxR binds with the non-naturally paired C12 to activate pLux, and promoter crosstalk, where LasR binds with C12 to activate non-naturally paired pLux. However, regulator crosstalk, in which the naturally paired autoinducer and promoter function through a cross-talking regulator protein, was not detected in this work.
When signal crosstalk is constructed and tested in the context of positive feedback, our results showed a significant shrinkage of the bistable region. Because of this topology’s bistable capability and wide presence in most bacterial QS decision-making circuits, such a decrease in bistability robustness due to QS crosstalk suggests a new strategy for developing anti-infection therapeutics. Namely, we might exploit “artificial” crosstalk to disrupt intercellular communication specificity and collapse the group’s coordination, which could be an efficient and economic approach in medical treatments, especially for QS-dependent bacterial infection.
On the other hand, promoter crosstalk caused complex trimodal responses when embedded within a positive feedback circuit. This can only be explained when network bistability, gene expression stochasticity, and genetic mutations are all taken into consideration. These results highlight the potential for engineering gene networks to express complex behaviors due to host-circuit interactions. We computationally predicted and experimentally verified that the C12-LasR-pLux positive feedback circuit could drive the formation of three subpopulations from an isogenic initial culture: one population expressing high GFP expression, the second showing basal GFP expression, and the third population with no GFP expression. The high and low GFP states are the result of positive feedback enabled bistability and gene expression stochasticity-induced random state transitions: commonly reported as a hallmark of many bistable systems (Acar et al., 2005; Gardner et al., 2000; Tan et al., 2009; Guido et al., 2006; Isaacs et al., 2003). This population heterogeneity is not caused by genetic factors.
The third non-GFP population is the result of genetic mutation from IS10 insertion. The mutation only happened in the C12-LasR-pLux positive feedback circuit but not in CP-LasR-pLux-C12 (Figure S 3D) or the C12-LuxR-pLux positive feedback circuit (Figure S 2B). It is, therefore, possible that the special sequence arrangements of the positive feedback circuit (for example, the symmetric pLux promoters flanking the LasR gene) on the plasmid coupled with the stress of exogenous protein overexpression led to transposon activation and gene network destruction. Given that many current synthetic gene circuits are constructed with a similar symmetric structure in a plasmid (such as Promoter-RBS-Gene1-RBS-Gene2-, or Promoter-RBS-Gene1-Terminator-Promoter-RBS-Gene2-Terminator), the mutation may occur for a wide range of engineered gene circuits. On the other hand, from an engineer’s perspective, the mutation stands in contrast to previously reported host-circuit interactions, which are primarily related to resource limitation and resulting growth defects (Brophy and Voigt, 2014). Here we were able to illustrate that both the components used and the topology of the network constructed could contribute to resource independent host-circuit interactions. This concept of combining nonlinear dynamics and host-circuit interactions to enrich population diversity expands our understanding of mechanisms contributing to cell-cell variability, and suggests new directions in engineering gene networks to utilize hybrid factors.
Taken together, our studies not only showcase living cells’ amazing complexity and the difficulty in the refining of engineered biological systems, but also reveal an overlooked mechanism by which multimodality arises from the combination of an engineered gene circuit and host-circuit interactions (Ellis et al., 2009; Hussain et al., 2014; Litcofsky et al., 2012; Nevozhay et al., 2013; Prindle et al., 2014).
SIGNIFICANCE
Widespread quorum-sensing (QS) enables bacteria to communicate and plays a critical role in controlling bacterial virulence. QS components have also been widely used in synthetic biology applications. However, effects of promiscuous QS crosstalk remain unexplored. Here we systematically studied the crosstalk between LuxR/I and LasR/I systems. Combining synthetic biology and mathematical modeling, this work reveals the complexity of QS crosstalk, which is critical for unraveling the underlying principles of bacterial decision-making and survival strategies for both natural and synthetic systems. Furthermore, the unusual hybrid multimodality arising from the combination of engineered gene circuits and circuit-host interactions could be utilized in biotechnology.
EXPERIMENTAL PROCEDURES
Strains, Growth Conditions and Media
All cloning experiments were performed in E.coli DH10B (Invitrogen, USA), and measurements of positive feedback response were conducted in DH10B and MG1655. Cells were grown at 37 °C (unless specified) in liquid and solid Luria-Bertani (LB) broth medium with 100 µg/mL ampicillin. Chemical 3OC6HSL and 3OC12HSL (Sigma-Aldrich, USA) were dissolved in ddH2O and DMSO, respectively. Cultures were shaken in 5 mL or 15 mL tubes at 220 rotations per minute (r.p.m), and inducers were added at OD600~0.1.
Plasmids Construction
Plasmids were constructed according to standard molecular cloning protocols and the genetic circuits were assembled using standardized BioBricks methods based on primary modules (Table S4) from the iGEM Registry (www.parts.igem.org). The receiver CP-LuxR-pLux was constructed from six BioBrick standard biological parts: BBa_K176009 (Constitutive promoter, CP), BBa_B0034 (Ribosome binding site, RBS), BBa_C0062 (luxR gene), BBa_B0015 (transcriptional terminator), BBa_R0062 (lux promoter), and BBa_E0240 (GFP generator, RBS-GFP-T). As an example, to produce the RBS-LuxR part, LuxR plasmid was digested by XbaI and PstI to produce a fragment while the RBS plasmid was digested by SpeI and PstI as the vector. The fragment and vector were purified by gel electrophoresis (1% TAE agarose gel) and extracted using a PureLink gel extraction kit (Invitrogen). Then, the fragment and vector were ligated together using T4 DNA ligase, the ligation products were transformed into E.coli DH10B and clones were screened by plating on 100 µg/mL ampicillin LB agar plates. Finally their plasmids were extracted and verified by double restriction digest (EcoRI and PstI) and DNA sequencing (Biodesign sequencing lab in ASU). After confirming that the newly assembled RBS-LuxR was correct, subsequent rounds to produce the RBS-LuxR-Terminator were performed similarly until completing the entire receiver CP-LuxR-pLux construction. All the other receivers and positive feedback circuits were assembled similarly. Restriction enzymes and T4 DNA ligase were from New England Biolabs. All the constructs were verified by sequencing step by step. To keep all the constructs’ expression consistent in the cell, we transferred all the fragments into the pSB1A3 vector before test.
Flow Cytometry
All the samples were analyzed at the time points indicated on an Accuri C6 flow cytometer (Becton Dickinson, USA) with 488 nm excitation and 530±15 nm emission detection (GFP). The data were collected in a linear scale and noncellular low-scatter noise was removed by thresholding. All measurements of gene expression were obtained from at least three independent experiments. For each culture, 20,000 events were collected at a slow flow rate. Data files were analyzed using MATLAB (MathWorks).
Hysteresis Experiment
For OFF→ON experiments, initially uninduced overnight culture was diluted into fresh media, grown at 37 °C and 220 r.p.m for about 1.5 h (OD600~0.1), then distributed evenly into new tubes and induced with various amounts of C6 or C12. Flow cytometry analyses were performed at 6, 12, and 21 hours to monitor the fluorescence levels, which generally became stable after 6 hours induction according to our experience. For ON→OFF experiments, initially uninduced cells were induced with 10−4 M (or 10−9 M) autoinducer and tested by flow cytometry to ensure they were fully induced. Cells were then collected with low-speed centrifugation, washed twice, resuspended with fresh medium, and at last inoculated into fresh medium with varying inducer concentrations at a 1:80 ratio. For the LasR-pLux positive feedback system, we only diluted once and grew them for 6, 12, 18, 24, or 32 hours, but for the other hysteresis experiments, the ON cells were collected and diluted twice into new medium with the same concentrations of C6 or C12 at 12 h and 24 h.
Growth Curve Assay
First, different initial states cells were collected: initial OFF cells were cells grown overnight without inducers, initial ON cells were initial OFF cells induced with 10−9 M C12 for 12 hours, and the Mutated cells were cells induced with 10−4 M C12 for 12 hours, diluted into fresh media with 10−4 M C12, and grown at 37 °C for another 12 h. Before the growth rate assay, all the cells’ fluorescence was tested by flow cytometry to verify their states. Growth rate was measured by using absorbance at 600 nm with a plate reader (BioTek, USA). Cells from each state were then diluted into fresh LB media (1000 µL, O.D. ~0.06) with 10−8 M C12 and grown at 37 or 34 °C. For each sample, OD was measured by using 200 µL cultures in a 96-well plate and tested over 24 hours. The experiments were independently replicated three times.
Microfludics, Fluorescence Microscopy, and Image Processing
The use of microfluidic devices coupled with fluorescence measurement allowed us to measure gene network dynamics in single cells. Media flow direction and speed was controlled through hydrostatic pressure. A detailed description of the chip can be found in the work of Ferry MS, et al (Ferry et al, 2011). Once the cell was loaded into the trap, the flow was reversed and its rate was slowed to ~120 µm/min to ensure that the cells would not be washed away and would receive enough nutrients. Furthermore, care was taken to avoid introducing bubbles to any part of the chip as they considerably disrupt flow. The chip temperature was maintained at 34 °C with an external microscope stage (Tokai Hit, Japan). Inducer concentrations were controlled by adjusting the heights of the inducer-containing media syringes relative to one another.
Images were taken using a Nikon Eclipse Ti inverted microscope (Nikon, Japan) equipped with an LED-based Lumencor SOLA SE Light Engine with the appropriate filter sets. The excitation wavelength for GFP was 472 nm, and fluorescence emission was detected with a Semrock 520/35 nm band pass filter. Phase and fluorescent images were taken under a magnification of 40×, and perfect focus was maintained automatically using Nikon Elements software.
Initially OFF cells (K-12 MG1655) induced with 10−9 M C12 (6 hours) were collected as the initial ON cells, washed, resuspended with fresh media and then loaded into the trap. 100 µg/mL ampicillin was added into media 1 and 2, but only media 2 was augmented with the corresponding inducer (10−8 M C12). The microfluidic device was used to control the chemical concentration by switching between media 1 and 2. For initial ON cells, media 2 was provided to the cells for the duration of the experiment. To prevent photobleaching and phototoxicity to the cells in the trap, exposure time was limited to 100 ms for GFP.
Images were taken every 5 minutes for about 28 hours in total. The pixels in all images are normalized to 0 – 1 range before analysis. One image was chosen for quantification every 15 minutes (i.e. three images). For each cell, the intensity was calculated by averaging three selected points (left, middle, and right) in the cell and then subtracting the background. Since all the cells are offspring of the first initial ON cell, each branch in Figure 5b stands for one progeny. The cells that were washed away or had less than three generations were not analyzed.
Mathematical Modeling
Ordinary differential equation models were solved and analyzed by MATLAB. Stochastic simulations were written in MATLAB and run on a standard personal computer (details are provided in Supplemental Information).
Supplementary Material
ACKNOWLEDGMENTS
We thank Jeff Hasty for the microfluidic setup and chip design. We also thank Riqi Su and Philippe Faucon for helpful discussions and suggestions. D.J.M is partially supported by ASU IRA Fulton School of Engineering’s Dean’s fellowship. This study was financially supported by National Science Foundation Grant DMS-1100309, American Heart Association Grant 11BGIA7440101, and National Institutes of Health Grant GM106081 (to X.W.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL INFORMATION
Supplemental Information contains full details about the mathematical modeling construction and parameter fittings, five figures and four tables and can be found with this article online.
AUTHOR CONTRIBUTIONS
X.W. and F.W. designed the study; F.W. performed the experiments and carried out the mathematical modeling; X.W. and F.W. analyzed the data; D.J.M. and F.W. made the microfluidic chips; F.W, D.J.M., and X.W. wrote the manuscript.
REFERENCES
- Acar M, Becskei A, van Oudenaarden A. Enhancement of cellular memory by reducing stochastic transitions. Nature. 2005;435:228–232. doi: 10.1038/nature03524. [DOI] [PubMed] [Google Scholar]
- Balagadde FK, Song H, Ozaki J, Collins CH, Barnet M, Arnold FH, Quake SR, You L. A synthetic Escherichia coli predator-prey ecosystem. Mol Syst Biol. 2008;4:187. doi: 10.1038/msb.2008.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu S, Mehreja R, Thiberge S, Chen M-T, Weiss R. Spatiotemporal control of gene expression with pulse-generating networks. Proc. Natl. Acad. Sci. U. S. A. 2004;101:6355–6360. doi: 10.1073/pnas.0307571101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu S, Gerchman Y, Collins CH, Arnold FH, Weiss R. A synthetic multicellular system for programmed pattern formation. Nature. 2005;434:1130–1134. doi: 10.1038/nature03461. [DOI] [PubMed] [Google Scholar]
- Brenner K, Karig DK, Weiss R, Arnold FH. Engineered bidirectional communication mediates a consensus in a microbial biofilm consortium. Proc. Natl. Acad. Sci. U. S. A. 2007;104:17300–17304. doi: 10.1073/pnas.0704256104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brophy JAN, Voigt CA. Principles of genetic circuit design. Nat. Methods. 2014;11:508–520. doi: 10.1038/nmeth.2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canton B, Labno A, Endy D. Refinement and standardization of synthetic biological parts and devices. Nat Biotechnol. 2008;26:787–793. doi: 10.1038/nbt1413. [DOI] [PubMed] [Google Scholar]
- Chen AY, Deng Z, Billings AN, Seker UOS, Lu MY, Citorik RJ, Zakeri B, Lu TK. Synthesis and patterning of tunable multiscale materials with engineered cells. Nat. Mater. 2014;13:515–523. doi: 10.1038/nmat3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang JS, Rivoire O, Leibler S. Simpson’s paradox in a synthetic microbial system. Science. 2009;323:272–275. doi: 10.1126/science.1166739. [DOI] [PubMed] [Google Scholar]
- Danino T, Mondragon-Palomino O, Tsimring L, Hasty J. A synchronized quorum of genetic clocks. Nature. 2010;463:326–330. doi: 10.1038/nature08753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis T, Wang X, Collins JJ. Diversity-based, model-guided construction of synthetic gene networks with predicted functions. Nat. Biotechnol. 2009;27:465–471. doi: 10.1038/nbt.1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferry MS, Razinkov IA, Hasty J. Microfluidics for synthetic biology: from design to execution. Methods Enzym. 2011;497:295–372. doi: 10.1016/B978-0-12-385075-1.00014-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuqua C, Winans SC, Greenberg EP. CENSUS AND CONSENSUS IN BACTERIAL ECOSYSTEMS: The LuxR-LuxI Family of Quorum-Sensing Transcriptional Regulators. Annu. Rev. Microbiol. 1996;50:727–751. doi: 10.1146/annurev.micro.50.1.727. [DOI] [PubMed] [Google Scholar]
- Gardner TS, Cantor CR, Collins JJ. Construction of a genetic toggle switch in Escherichia coli. Nature. 2000;403:339–342. doi: 10.1038/35002131. [DOI] [PubMed] [Google Scholar]
- Gillespie D. Exact stochastic simulation of coupled chemical reactions. J Phys Chem. 1977;81:2340–2361. [Google Scholar]
- Gray KM, Passador L, Iglewski BH, Greenberg EP. Interchangeability and specificity of components from the quorum-sensing regulatory systems of Vibrio fischeri and Pseudomonas aeruginosa. J. Bacteriol. 1994;176:3076–3080. doi: 10.1128/jb.176.10.3076-3080.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guido NJ, Wang X, Adalsteinsson D, McMillen D, Hasty J, Cantor CR, Elston TC, Collins JJ. A bottom-up approach to gene regulation. Nature. 2006;439:856–860. doi: 10.1038/nature04473. [DOI] [PubMed] [Google Scholar]
- Hong SH, Hegde M, Kim J, Wang X, Jayaraman A, Wood TK. Synthetic quorum-sensing circuit to control consortial biofilm formation and dispersal in a microfluidic device. Nat. Commun. 2012;3:613. doi: 10.1038/ncomms1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain F, Gupta C, Hirning AJ, Ott W, Matthews KS, Josić K, Bennett MR. Engineered temperature compensation in a synthetic genetic clock. Proc. Natl. Acad. Sci. 2014:201316298. doi: 10.1073/pnas.1316298111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaacs FJ, Hasty J, Cantor CR, Collins JJ. Prediction and measurement of an autoregulatory genetic module. Proc. Natl. Acad. Sci. 2003;100:7714–7719. doi: 10.1073/pnas.1332628100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayaraman A, Wood TK. Bacterial quorum sensing: signals, circuits, and implications for biofilms and disease. Annu. Rev. Biomed. Eng. 2008;10:145–167. doi: 10.1146/annurev.bioeng.10.061807.160536. [DOI] [PubMed] [Google Scholar]
- Ji G, Beavis RC, Novick RP. Cell density control of staphylococcal virulence mediated by an octapeptide pheromone. Proc. Natl. Acad. Sci. U. S. A. 1995;92:12055–12059. doi: 10.1073/pnas.92.26.12055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan HB, Greenberg EP. Diffusion of autoinducer is involved in regulation of the Vibrio fischeri luminescence system. J. Bacteriol. 1985;163:1210–1214. doi: 10.1128/jb.163.3.1210-1214.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Kievit TR, Iglewski BH. Bacterial quorum sensing in pathogenic relationships. Infect. Immun. 2000;68:4839–4849. doi: 10.1128/iai.68.9.4839-4849.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi H, Kaern M, Araki M, Chung K, Gardner TS, Cantor CR, Collins JJ. Programmable cells: interfacing natural and engineered gene networks. Proc Natl Acad Sci U A. 2004;101:8414–8419. doi: 10.1073/pnas.0402940101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovarík A, Matzke MA, Matzke AJ, Koulaková B. Transposition of IS10 from the host Escherichia coli genome to a plasmid may lead to cloning artefacts. Mol. Genet. Genomics MGG. 2001;266:216–222. doi: 10.1007/s004380100542. [DOI] [PubMed] [Google Scholar]
- LaSarre B, Federle MJ. Exploiting Quorum Sensing To Confuse Bacterial Pathogens. Microbiol. Mol. Biol. Rev. 2013;77:73–111. doi: 10.1128/MMBR.00046-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litcofsky KD, Afeyan RB, Krom RJ, Khalil AS, Collins JJ. Iterative plug-and-play methodology for constructing and modifying synthetic gene networks. Nat. Methods. 2012;9:1077–1080. doi: 10.1038/nmeth.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B. The significance of responses of the genome to challenge. Science. 1984;226:792–801. doi: 10.1126/science.15739260. [DOI] [PubMed] [Google Scholar]
- Meighen EA. Genetics of Bacterial Bioluminescence. Annu. Rev. Genet. 1994;28:117–139. doi: 10.1146/annurev.ge.28.120194.001001. [DOI] [PubMed] [Google Scholar]
- Miller MB, Bassler BL. Quorum Sensing in Bacteria. Annu. Rev. Microbiol. 2001;55:165–199. doi: 10.1146/annurev.micro.55.1.165. [DOI] [PubMed] [Google Scholar]
- Nevozhay D, Zal T, Balazsi G. Transferring a synthetic gene circuit from yeast to mammalian cells. Nat. Commun. 2013;4:1451. doi: 10.1038/ncomms2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng W-L, Bassler BL. Bacterial Quorum-Sensing Network Architectures. Annu. Rev. Genet. 2009;43:197–222. doi: 10.1146/annurev-genet-102108-134304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsubo Y, Genka H, Komatsu H, Nagata Y, Tsuda M. High-temperature-induced transposition of insertion elements in burkholderia multivorans ATCC 17616. Appl. Environ. Microbiol. 2005;71:1822–1828. doi: 10.1128/AEM.71.4.1822-1828.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozbudak EM, Thattai M, Lim HN, Shraiman BI, Van Oudenaarden A. Multistability in the lactose utilization network of Escherichia coli. Nature. 2004;427:737–740. doi: 10.1038/nature02298. [DOI] [PubMed] [Google Scholar]
- Pai A, Tanouchi Y, You L. Optimality and robustness in quorum sensing (QS)-mediated regulation of a costly public good enzyme. Proc. Natl. Acad. Sci. U. S. A. 2012;109:19810–19815. doi: 10.1073/pnas.1211072109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne S, Li B, Cao Y, Schaeffer D, Ryser MD, You L. Temporal control of self-organized pattern formation without morphogen gradients in bacteria. Mol. Syst. Biol. 2013;9 doi: 10.1038/msb.2013.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez PD, Weiss JT, Hagen SJ. Noise and crosstalk in two quorum-sensing inputs of Vibrio fischeri. BMC Syst. Biol. 2011;5:153. doi: 10.1186/1752-0509-5-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova EV, Håvarstein LS, Morrison DA. Regulation of competence for genetic transformation in Streptococcus pneumoniae by an auto-induced peptide pheromone and a two-component regulatory system. Mol. Microbiol. 1996;21:853–862. doi: 10.1046/j.1365-2958.1996.501417.x. [DOI] [PubMed] [Google Scholar]
- Piper KR, Beck von Bodman S, Farrand SK. Conjugation factor of Agrobacterium tumefaciens regulates Ti plasmid transfer by autoinduction. Nature. 1993;362:448–450. doi: 10.1038/362448a0. [DOI] [PubMed] [Google Scholar]
- Prindle A, Samayoa P, Razinkov I, Danino T, Tsimring LS, Hasty J. A sensing array of radically coupled genetic /`biopixels/’. Nature. 2012;481:39–44. doi: 10.1038/nature10722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prindle A, Selimkhanov J, Li H, Razinkov I, Tsimring LS, Hasty J. Rapid and tunable post-translational coupling of genetic circuits. Nature. 2014;508:387–391. doi: 10.1038/nature13238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster M, Urbanowski ML, Greenberg EP. Promoter specificity in Pseudomonas aeruginosa quorum sensing revealed by DNA binding of purified LasR. Proc. Natl. Acad. Sci. U. S. A. 2004;101:15833–15839. doi: 10.1073/pnas.0407229101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seed PC, Passador L, Iglewski BH. Activation of the Pseudomonas aeruginosa lasI gene by LasR and the Pseudomonas autoinducer PAI: an autoinduction regulatory hierarchy. J. Bacteriol. 1995;177:654–659. doi: 10.1128/jb.177.3.654-659.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solano C, Echeverz M, Lasa I. Biofilm dispersion and quorum sensing. Curr. Opin. Microbiol. 2014;18:96–104. doi: 10.1016/j.mib.2014.02.008. [DOI] [PubMed] [Google Scholar]
- Sousa A, Bourgard C, Wahl LM, Gordo I. Rates of transposition in Escherichia coli. Biol. Lett. 2013;9:20130838. doi: 10.1098/rsbl.2013.0838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens AM, Greenberg EP. Quorum sensing in Vibrio fischeri: essential elements for activation of the luminescence genes. J. Bacteriol. 1997;179:557–562. doi: 10.1128/jb.179.2.557-562.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor JJ, Salis HM, Simpson ZB, Chevalier AA, Levskaya A, Marcotte EM, Voigt CA, Ellington AD. A synthetic genetic edge detection program. Cell. 2009;137:1272–1281. doi: 10.1016/j.cell.2009.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamsir A, Tabor JJ, Voigt CA. Robust multicellular computing using genetically encoded NOR gates and chemical /`wires/’. Nature. 2011;469:212–215. doi: 10.1038/nature09565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan C, Marguet P, You L. Emergent bistability by a growth-modulating positive feedback circuit. Nat. Chem. Biol. 2009;5:842–848. doi: 10.1038/nchembio.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzer K, Falconer C, Garber NC, Diggle SP, Camara M, Williams P. The Pseudomonas aeruginosa lectins PA-IL and PA-IIL are controlled by quorum sensing and by RpoS. J. Bacteriol. 2000;182:6401–6411. doi: 10.1128/jb.182.22.6401-6411.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M, Su R-Q, Li X, Ellis T, Lai Y-C, Wang X. Engineering of regulated stochastic cell fate determination. Proc. Natl. Acad. Sci. 2013;110:10610–10615. doi: 10.1073/pnas.1305423110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong W, Ferrell JE. A positive-feedback-based bistable ’memory module” that governs a cell fate decision. Nature. 2003;426:460–465. doi: 10.1038/nature02089. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.