Abstract
We characterized two newly isolated myoviruses, Bp8p-C and Bp8p-T, infecting the ginger rhizome rot disease pathogen Bacillus pumilus GR8. The plaque of Bp8p-T exhibited a clear center with a turbid rim, suggesting that Bp8p-T could transform into latent phage. Lysogeny assays showed that both the two phages could form latent states, while Bp8p-T could form latent phage at a higher frequency and stability than Bp8p-C. The genomes of Bp8p-C and Bp8p-T were 151,417 and 151,419 bp, respectively; both encoded 212 putative proteins, and only differed by three nucleotides. Moreover, owing to this difference, Bp8p-C encoded a truncated, putative actin-like plasmid segregation protein Gp27-C. Functional analysis of protein Gp27 showed that Gp27-T encoded by Bp8p-T exhibited higher ATPase activity and assembly ability than Gp27-C. The results indicate that the difference in Gp27 affected the phage lysogenic ability. Structural proteome analysis of Bp8p-C virion resulted in the identification of 14 structural proteins, among which a pectin lyase-like protein, a putative poly-gamma-glutamate hydrolase, and three proteins with unknown function, were firstly identified as components of the phage virion. Both phages exhibited specific lytic ability to the host strain GR8. Bp8p-C showed better control effect on the pathogen in ginger rhizome slices than Bp8p-T, suggesting that Bp8p-C has a potential application in bio-control of ginger rhizome rot disease.
INTRODUCTION
Bacteriophages (phages) are viral predators of bacteria that propagate by infecting the host (1) and, based on the mode of replication of the phage genome, they are classified as lytic or lysogenic phages. To maintain the lysogenic state, phage gene expression is regulated by the phage lysogenic regulatory system. The lytic switch mechanism of the temperate phage λ has been well studied (2), which illustrates the classical lysogen conversion mechanism of λ and similar phages. Several other strategies for phage lysogenic conversion have also been observed. For example, a single nucleotide mutation in the LexA-binding site of phage SPC32H induces phage SPC32H to maintain the lysogenic state (1). According Yoshihiko et al., a temperate phage, c-st, with a linear genome can transform into a circular plasmid prophage in the host, Clostridium botulinum, which provides phage c-st the ability to exhibit unstable lysogeny (3). Meanwhile, the TubZ-like plasmid segregation system protein encoded by c-st is essential for partition of the prophage plasmid (4).
Plasmid segregation systems are essential for accurate partition of large and essential plasmids to daughter cells during cell division (5). The plasmid segregation system typically consists of three components: a centromere-like DNA region, a small DNA-binding adaptor protein, and a nucleotide-driven motor protein with NTPase activity. Based on the nature of the motor proteins, three major classes of segregation systems have been identified to date and can be described as ParA-like system (type I, ParABS system), ParM-like system (type II, ParMRC system), and TubZ-like system (type III). Among the three classes of plasmid segregation systems described above, type III segregation system has been studied in detail in phage c-st (4). A tubulin protein encoded by phage 201ϕ2-1 can form a spindle-like array in vivo, which facilitates the phage genome to locate the center of the cell and optimizes phage reproduction (6). The ParMRC system is currently the most well-characterized plasmid segregation system and has been identified in numerous plasmids in Gram-positive and Gram-negative bacteria (7); however, to our knowledge, no ParMRC system has been characterized in phages to date.
Bacillus pumilus, a Gram-positive bacterium belonging to the B. subtilis group (8), has been widely used as a plant growth-promoting rhizobacterium to prevent plant disease caused by pathogenic bacteria and fungi (9). However, numerous B. pumilus strains have also been reported to cause several human and plant diseases (10, 11). B. pumilus strain, such as MB2 and MB4, isolated from Egypt, can cause leaf blight of mango trees (12), and another B. pumilus strain was found to be a pathogen of Asian pear tree leaf and twig dieback (13). B. pumilus also causes some human disease (10), such as acting as a bloodstream pathogen during infancy (11). B. pumilus strain GR8 isolated from Shandong Province of China was identified as an opportunistic pathogen of the ginger rhizome rot disease (14) and causes serious losses of ginger production in Shandong Province. Due to the ability of phages and phage-derived endolysin in controlling bacterial pathogens, phages are regarded as potential agents for plant disease control. Before 1981, more than 33 different phages were reported to infect B. pumilus, and a few more have been characterized since (15). In addition, the genome of 10 B. pumilus phages have been characterized to date (16, 17); however, the structural proteome of the phage virion has not yet been analyzed.
In this study, we characterized two newly isolated myoviruses, Bp8p-C and Bp8p-T, infecting B. pumilus GR8, the pathogen causing ginger rhizome rot disease (14), by physiological analysis, genome sequencing, and structural proteomic analysis. Further, we clarified the effect of actin-like plasmid segregation-associated protein on phage lysogeny and revealed a direct relationship between the actin-like protein of the plasmid segregation system and the lysogenic ability of phage.
MATERIALS AND METHODS
Phage isolation and genome DNA purification.
B. pumilus strain GR8, a ginger rhizome rot-causing pathogen, was isolated at our laboratory (14) and was cultured in Luria-Bertani (LB) broth at 30°C. Soil samples were collected from Shandong Province, China, where ginger rhizome rot disease is prevalent. Phage isolation, purification, and the efficiency-of-plating test were carried out by double-agar overlay assay as previously described (18) with some modifications. B. pumilus strain GR8 was incubated overnight, and a 0.1-g soil sample or rotted ginger rhizome sample was added to the culture. After incubation for 8 h, the supernatant was collected by centrifugation at 8,000 × g at 4°C for 30 min and filtered through a 0.22-μm-pore-size filter. Next, a 100-μl sample, 200 μl of log-phase B. pumilus GR8, and 4 ml of semisolid LB medium were mixed and overlaid onto an LB agar plate. After several rounds of plaque purification, a phage sample forming consistent plaques was regarded as pure. The phage obtained was propagated by washing off the plaques formed on the semisolid plate with SM buffer (0.58% [wt/vol] NaCl, 0.2% [wt/vol] Mg2SO4, 50 mM Tris-HCl [pH 7.5]). The phage genome was purified by phenol-chloroform extraction with protease K-sodium dodecyl sulfate (SDS) treatment (19). For other analyses, the phage was purified by sucrose density gradient centrifugation as previously described (20).
Electron microscopy.
The purified phage suspension was deposited on cuprum grids with carbon-coated Formvar film and stained with 2% potassium phosphotungstate (pH 7.2) (21). After air drying, the sample was observed using a transmission electron microscope (H-7000FA; Hitachi, Tokyo, Japan) at an acceleration voltage of 100 kV.
One-step growth curve.
A one-step growth curve and burst size assay were carried out as previously described, with some modifications (22). The phage was incubated with log-phase B. pumilus GR8 at a multiplicity of infection (MOI) of 1.0, at 30°C for 30 min for phage adsorption. The nonadsorbed phage was removed by centrifugation at 10,000 × g for 1 min, and the pellet was resuspended in 50 ml of LB broth. The phage titer in the culture was determined every 30 min. The phage burst size was determined, as previously described (22). To analyze the effect of different MOIs on bacterial growth, B. pumilus GR8 was infected with phage at different MOIs (0.01, 0.1, 1.0, and 10), and the absorbance of the culture was determined at 600 nm every 30 min. All experiments were carried out in triplicate.
Phage stability assay during storage.
The purified phage was stored at −80, 4, and 30°C, and the phage titer was determined by double-agar overlay assay immediately and every week after storage.
Phage genome sequencing and bioinformatics analysis.
The ends of the phage genomes were analyzed by digestion with exonuclease III and BAL 31 nuclease (TaKaRa, Shiga, Japan) (23). The genome was sequenced using a GS FLX system (Roche, Basel, Switzerland) and assembled into contigs using a GS de novo Assembler (Roche). The remaining gaps between the contigs of the phage genomes were filled-in via primer walking. The coding sequences (CDSs) of the phage genomes were predicted using the FGENE SV software (Softberry) and by visual inspection. The putative function of each gene was predicted by performing a search in the National Center for Biotechnology Information nonredundant (NR) and CDD databases (24) using the basic local alignment search tool (BLAST). The motif and functional domain composition of the predicted protein were analyzed by searching the Pfam database (25) and by using HHpred (26). Phage genome annotation was visualized by using CGview (27). The COG (Cluster of Orthologous Groups) classifications of the proteins encoded by the phages were classified by using COGnitor (28). The genes encoding the putative tRNAs were analyzed using tRNAScan (29). Tandem repeat and insert sequences in the phage genome were analyzed by using Tandem Repeat Finder (30) and ISfinder (31), respectively. Comparative genome analysis of the phages was carried out using Gepard (32), Mauve (33), EMBOSS Stretcher (http://emboss.bioinformatics.nl/cgi-bin/emboss/stretcher), and CoreGenes 3.0 (34). Genes with scores above 75 were regarded as core genes. The phylogenetic trees in the present study were constructed using Mega 5.0 (35).
Lysogeny assay.
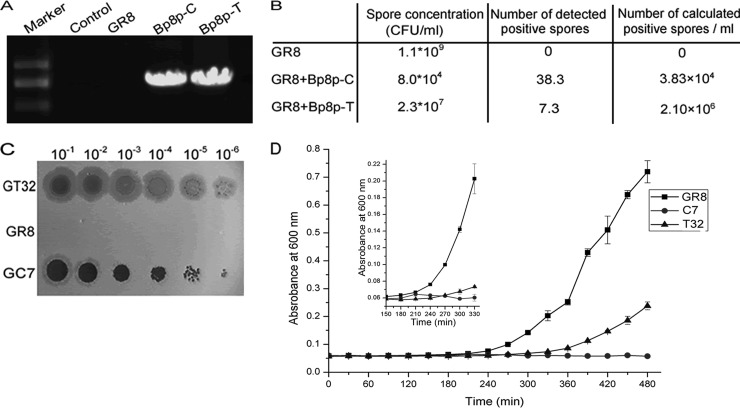
To evaluate whether the phage could convert to the latent state, B. pumilus strain GR8 was infected with the phage at an MOI of 1.0. After incubation for 96 h, the culture was boiled at 85°C for 30 min to remove vegetative cells and phage (36). The spore concentration was determination by agar plate dilution method, and the positive spores containing latent phage was identified by amplification of the gene orf27 in the phage genomes.
Expression, purification, and functional analyses of gene orf27.
DNA manipulation was performed as previously described (37). Gene orf27 from both phages, Bp8p-C and Bp8p-T, were amplified and inserted into plasmid pET28a to construct recombinant plasmids. The primers used were ORF27T-F/BamHI (5′-TAGGATCCATGTGCCCGGGGAGCTAC-3′) and ORF27-R/SalI (5′-ATGTCGACCTAGCCCCCATAATGATCAGC-3′) to amplify orf27-T from phage Bp8p-T and ORF27C-F/BamHI (5′-TGGGATCCATGATCACAGTAGAGCATG-3′) and ORF27-R/SalI (5′-ATGTCGACCTAGCCCCCATAATGATCAGC-3′) to amplify orf27-C from phage Bp8p-C. The recombinant plasmids were transformed into Escherichia coli strain BL21 to produce recombinant E. coli strains. The proteins were expressed by induction with 0.4 mM IPTG (isopropyl-β-d-thiogalactopyranoside) and the protein was purified by nickel-nitrilotriacetic acid chromatography (Ni-NTA columns; Qiagen, Dusseldorf, Germany) and buffer exchanged into 25 mM Tris (pH 7.5), 300 mM NaCl, 5% glycerol, and 1 mM β-mercaptoethanol (38). The ATPase activity of the proteins was determined by using an phosphate assay kit (E-6646) from Life Technologies (Carlsbad, CA) according to the manufacturer's instructions. The polymerization ability of the proteins was determined by analyzing the precipitation of the proteins as previously described (39). Briefly, MgCl2 and ATP (Sigma, Milpitas, CA) were added to the protein to final concentrations of 5 and 2 mM, respectively. After incubation at 30°C for 30 min and centrifugation at 10,000 × g for 20 min, the supernatant and pellet fractions were separated by SDS–15% PAGE. A Western blot was carried out with the protein using mouse monoclonal antibody anti-His6 (2) (Roche) as previously described (40). ATPase activity assay and determination of polymerization ability were carried out in triplicate.
Proteome analysis.
Phage purified by source density gradient centrifugation was resuspended in loading buffer (1% [wt/vol] SDS, 6% [wt/vol] sucrose, 100 mM 1,4-dithiothreitol, 10 mM Tris-HCl [pH 6.8], 0.0625% [wt/vol] bromophenol blue) and boiled at 90°C for 3 min (41). After separation by SDS–12% PAGE, the gel was stained with Coomassie blue and destained with water. A single visible band on the gel was excised, and the protein was digested in situ with trypsin, followed by peptide elution, chromatography, and MALDI-TOF-TOF MS (matrix-assisted laser desorption ionization–tandem time of flight mass spectrometry) on an Ultraflex MALDI-TOF-TOF mass spectrometer (Bruker, Karlsruhe, Germany) as described previously (42). The MS data were analyzed using Mascot software, version 2.3.01, and Sequest software, version 1.2.0.208, against a local database of all possible phage proteins.
Phage therapy for ginger rhizome rot disease caused by B. pumilus infection.
To study the efficiency of phages to control ginger rhizome rot disease caused by B. pumilus GR8, each phage was tested at a concentration range from 104 to 109 PFU/ml. The LaiWu Big ginger rhizome initiated from sterile tissue culture was cut into 3-mm slices and placed onto sterile, moistened filter-paper in a petri dish and inoculated with 100 μl of 108 CFU/ml, log-phase, bacterial suspension along with 100 μl of phage suspension at different concentrations. After incubation at 30°C for 24 h, disease severity ratings were evaluated as previous standards, where 1 denotes healthy tissue, 2 denotes slight (up to 20%) rot (slight discoloration), 3 denotes moderate (up to 50%) rot (discoloration and tissue breakdown), 4 denotes severe rot (50 to 75% of slice is affected), and 5 denotes complete rot (up to the entire slice is affected) (14). Sterile water served as negative control, while bacterial suspension alone added to the ginger rhizome slice was used as positive control. For each treatment, five ginger rhizome slices were used, and the experiment was carried out in triplicate.
Nucleotide sequence accession numbers.
The genome of phages Bp8p-C and Bp8p-T were deposited in GenBank database under accession numbers KJ010547 and KJ010548, respectively.
RESULTS
Isolation and identification of B. pumilus phages.
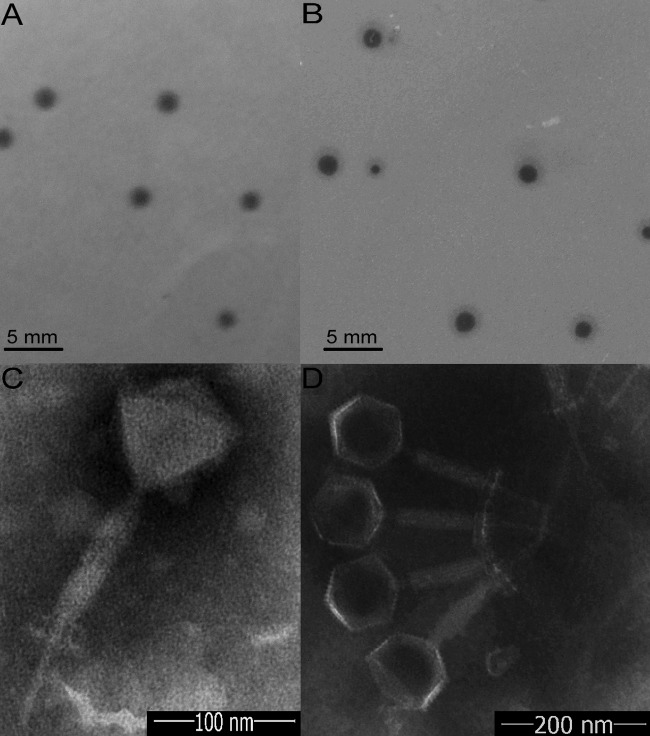
Five samples of rot ginger rhizomes and nine samples of surrounding soil were collected from Shandong Province of China to isolate B. pumilus GR8 phage. No phages were obtained from the rot ginger rhizome samples in the present study; however, two phages with different plaque morphology were isolated from the same soil sample. Of these, one phage, named Bp8p-C, formed relative clear, 2-mm plaques (Fig. 1A), while the other phage, named Bp8p-T, formed plaques with clear center of ∼2 mm and a turbid rim (Fig. 1B). The plaque features of phage Bp8p-T suggested that the phage might be temperate phage.
FIG 1.
Morphology of phage plaques and virions. Morphology analyses show the morphology of plaques formed by Bp8p-C (A) and Bp8p-T (B) on double-agar overlay plates and the virion morphology of phages Bp8p-C (C) and Bp8p-T (D). The phage virions were stained with potassium phosphotungstate and observed using transmission electron microscopy.
Based on the morphology of the two phages examined by transmission electron microscopy (Fig. 1C and D), Bp8p-C and Bp8p-T were classified as members of the Myoviridae family. Each of the two phages had an isometric head with a diameter of 88 nm, a 189-nm-long contractile tail, a 60-nm-long tail needle, and a 20-nm-long and 46-nm-wide baseplate. To date, a few phages of B. pumilus have been isolated (17), while only one B. pumilus phage from the Myoviridae family has been morphologically characterized (16).
One-step growth curve and infective activity of phages Bp8p-C and Bp8p-T.
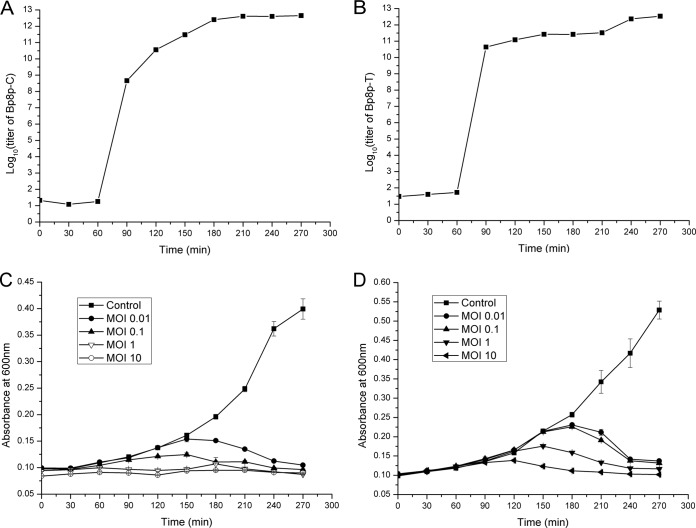
The one-step growth curve and infective activity of phage Bp8p-C and Bp8p-T were determined (Fig. 2). For Bp8p-C, the latent period was about 60 min, and calculated burst size was about 91, which was higher than that of other phages proposed for bacterial pathogen control (23), but lower than that of the B. pumilus phage phiAGATE. In addition, the multiplication period reached a plateau at about 180 min (Fig. 2A). For Bp8p-T, the latent period was also about 60 min as well, while the calculated burst size was 11, which was significantly lower than that of Bp8p-C. The phage titer of Bp8p-T after 240 min of infection was approximately equal to that of Bp8p-C at 180 min after infection, suggesting that the lysis of host strain infected by Bp8p-T might be delayed (Fig. 2B). The infective activity assay showed that the growth of host bacterium was obviously inhibited with an increase in phage concentration, even at the lowest MOI (Fig. 2C and D) for both phages. B. pumilus GR8 showed scarce growth when the MOI values of Bp8p-C and Bp8p-T were higher than 1.0 and 10, respectively. In addition, the optical density of the bacterial cultures decreased at 150 min after phage infection due to cell lysis triggered by phage release.
FIG 2.
One-step growth curve and infective activity of phages Bp8p-C and Bp8p-T. One-step growth curves of phages Bp8p-C (A) and Bp8p-T (B) were determined. The infective activity of phages Bp8p-C (C) and Bp8p-T (D) on host strain B. pumilus GR8 was determined.
Host range and storage stability of phages Bp8p-C and Bp8p-T.
The host range of the two phages was tested on B. thuringiensis strain CS-33, B. subtilis strain 168, B. cereus strain 411A, and B. anthracis strain 63605 (19), in addition to 14 different B. pumilus strains. The results showed that phages Bp8p-C and Bp8p-T could only infect 7 of the 14 B. pumilus strains tested and exhibited the same host range (see Table S1 in supplemental material). Neither phage could infect strains from the other bacterial species. Due to this specificity of Bp8p-C and Bp8p-T to infect B. pumilus, they exhibit potential as agents for controlling ginger rhizome rot disease. A comparison of the properties between phage Bp8p-C and Bp8p-T is shown in Table 1.
TABLE 1.
Comparison of the characteristics between phage Bp8p-C and Bp8p-T
| Phage name | Plaque morphology | Host range | Lysogenic ability | Genome size (bp) | CDS no. | Gp27 |
||
|---|---|---|---|---|---|---|---|---|
| Length (aa)a | ATPase activity | Assembly ability | ||||||
| Bp8p-C | Clear | 7 B. pumilus isolates | Low | 151,417 | 212 | 197 | Low | Low |
| Bp8p-T | Clear center, turbid rim | 7 B. pumilus isolates | High | 151,419 | 212 | 248 | High | High |
aa, amino acids.
The storage stability assay revealed that phage Bp8p-C and Bp8p-T was stable at 30°C for up to 14 days of storage, but the titer quickly decreased thereafter. Although the stability at 30°C was low, these two phages could maintain a high titer at 4°C after 4 months of storage, and >90% of the phage exhibited infective activity after 4 months of storage at −80°C.
Genomic characteristics of two new B. pumilus phages.
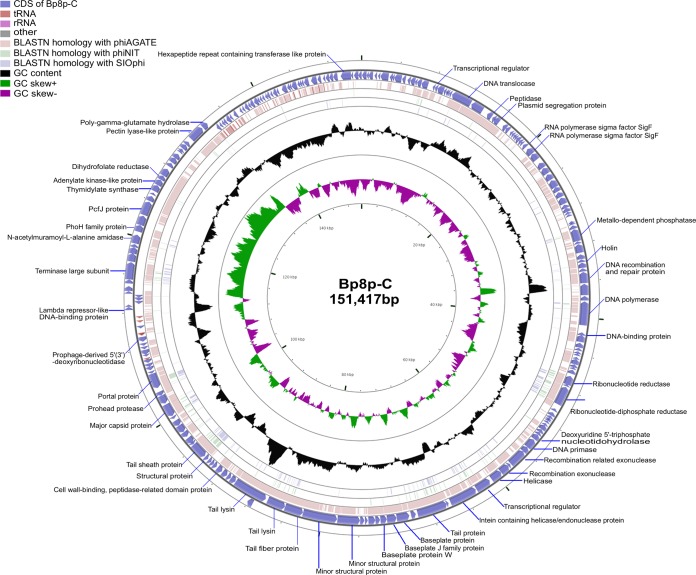
Both phages Bp8p-C and Bp8p-T showed a circular genome with a G+C content of 41.15%, and the genome sizes were 151,417 and 151,419 bp, respectively. Genome analysis revealed that the genomes of the two phages were highly similar, except for two guanine nucleotides inserted at positions 15999 to 16000 and one mutation causing a nucleotide change from thymine to cytosine at position 148412 in the Bp8p-T genome. A comparison of the genome characteristics between phage Bp8p-C and Bp8p-T is shown in Table 1. Considering their high similarity, we have described the genome characteristics of phage Bp8p-C only in detail. The genome of phage Bp8p-C had 212 putative coding sequences (CDSs) with an average length of 614 bp (Fig. 3; see Table S2 in supplemental material). The average distance between consecutive CDSs was 102.8 bp. Most of the CDSs were transcribed in reverse, and 29 were transcribed in the forward direction, in which 26 were clustered. A comprehensive search of the NR database for homologs of the 212 CDSs returned 180 significant matches (E-value ≤ 10−3), in which 44 CDSs only exhibited similarity to the genes of B. pumilus Myovirus phiAGATE. ORFans is a cluster of genes that are found in the genome of only one specific organism or showing orthology with genes from a closely related organism (43). We found 76 genes (35.8% of CDSs) that were specific for Bp8p-C or only homologous with genes of phiAGATE, which could be classified as ORFans and might provide Bp8p-C with unique features.
FIG 3.
Genome structure and comparative genome analysis of Bp8p-C. The outermost ring presents the CDSs of the circular Bp8p-C genome (blue) and the adjacent two rings shows BLASTN homology between Bp8p-C and phiAGATE (pink), as well as between Bp8p-C and phiNIT (light green). The middle ring indicates the G+C content (black), and the innermost ring represents the GC skew of Bp8p-C genome. The predicted functions of the CDSs are indicated.
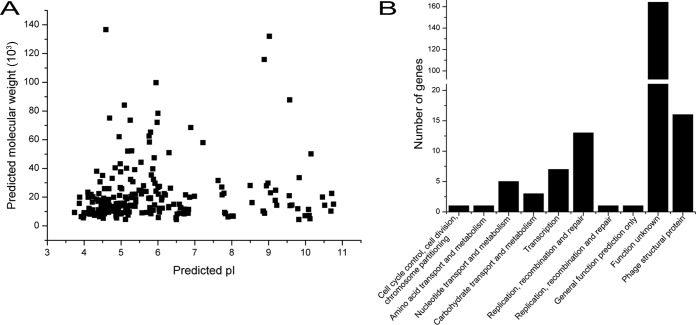
To determine whether the proteins encoded by Bp8p-C genome fit with the intracellular environment of the host strain, the predicted molecular mass and isoelectric point (pI) of the putative proteins encoded by Bp8p-C genome were analyzed (Fig. 4A). The results showed that most of the putative proteins (72.6% of the encoded CDSs) maintained a pI range between 4.0 and 7.0 with a molecular mass of ≤60 kDa. Proteomic analysis of B. pumilus revealed that B. pumilus intracellular proteins mainly exhibited a pI range from 4.0 to 7.0 (44). The distribution of theoretical phage protein pI fitted with the intracellular environment of the host strain, which was critical for phage infection and propagation. COG classification of the predicted phage proteins was performed (Fig. 4B). Besides the proteins with unknown functions, the majority of the annotated proteins were associated with phage structure (16 CDSs), DNA replication, recombination and repair (14 CDSs), and transcription (7 CDSs), as well as nucleotide transport and metabolism (5 CDSs). In addition, 19 putative proteins (9.0% of the encoded CDSs) were involved in DNA processing (including replication, repair, transcription, and nucleotide metabolism). ORF172 in the Bp8p-C genome was annotated to encode a pectin lyase-like protein, which could lyse the cell wall of the plant and was associated with pathogenicity of the plant bacterial pathogen (45). However, a previous report has indicated that a protein similar to Gp172 encoded by phages might disrupt bacterial biofilms and increase infective activity of the phage in bacteria (46). Based on these studies, subsequent experiments need to be conducted to evaluate the safety of phage-mediated plant pathogen control. The host lysis associated protein-encoding genes (endolysin-encoding gene orf155 and holin gene orf58), which are usually clustered together and synergistically lyse the host cell wall to release mature phages (47), were scattered in the Bp8p-C genome.
FIG 4.
Features of the proteome from Bp8p-C. (A) Theoretical molecular weight (Mw) and pI of putative phage proteins calculated by using the tool Compute pI/Mw. Each point indicates the predicted pI and Mw of a single protein. (B) COG classifications of the predicted proteins encoded by Bp8p-C genome. The phage structural associated proteins are indicated as phage structural protein.
Phage Bp8p-C contained more tRNAs than some other phages from the Myoviridae family (48). Five tRNAs, encoding tRNAMet, tRNAHis, tRNAGly, tRNAPhe, and tRNAAsn were found in the intergenic region of the Bp8p-C genome, while 2 of these overlapped each other (Fig. 3; see Table S3 in supplemental material). In addition, eight insertion sequences, which belonged to the families of IS3, IS4, and Tn3, as well as five tandem repeat sequences were also found in the Bp8p-C genome (see Tables S4 and S5 in supplemental material).
Comparative genome analysis.
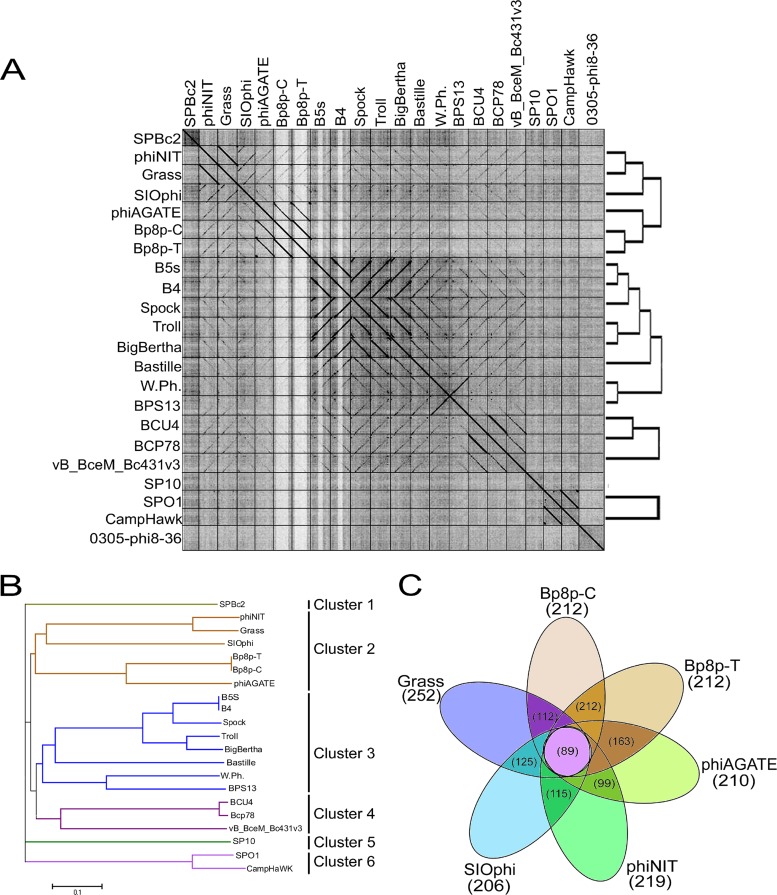
The genome similarities of the Myoviridae phages from the Bacillus genus were analyzed, and the results showed that phage Bp8p-C only exhibited high similarity to phage Bp8p-T (99.99%) and phiAGATE (62.5%). The genome of phage Bp8p-C also exhibited similarity to phage SIOphi (47.8%), phiNIT (47.4%), and Grass (47.4%), while the similarity to other Bacillus phages was low (Fig. 5A). A long terminal repeat sequence was found in the genome of phage Bp8p-C and phiAGATE by dot plot analysis of the genomes (see Fig. S1 in supplemental material).
FIG 5.
Comparative genome analysis of phage Bp8p-C. (A) Dot plot analysis of the Myoviridae family phages genome from Bacillus phages prepared using Gepard. The genome sequences of 22 Bacillus phages are used, and the locations of phage genomes are indicated on each axis. The linearity regions between the phage genomes are showed as a black line. (B) Phage phylogenetic tree based on the whole genome sequences of 21 Myoviridae phages of Bacillus. Phages belonging to different clusters are indicated. (C) Core-genome analysis of the six phages belonging to cluster 2. The numbers of core genes for the six phages are indicated in the central circle, and the numbers of core genes between adjacent phages are indicated in the cross regions.
To analyze the evolutionary relationship, a phylogenetic tree based on the whole genome sequence of 21 Myoviridae phages from the Bacillus genus was constructed (Fig. 5B). The phylogenetic tree revealed that Myoviridae phages of the Bacillus genus could be grouped into six clusters. Among these, phage Bp8p-C, Bp8p-T, phiAGATE, SIOphi, phiNIT, and Grass were grouped together as cluster 2. The dot plot and phylogenetic tree results were consistent. The phages in cluster 2 exhibited higher similarity than those in other clusters.
The proteomes of cluster 2 phages were analyzed using CoreGenes 3.0 software, and 89 proteins were found to be conserved in all six phages of cluster 2 (Fig. 5C). The functions of these proteins were mainly related to phage structure and DNA replication and repair, as well as host lysis, which are essential to the phage life cycle. Although phage Bp8p-C and phiAGATE exhibited high genome similarity at 62.5% and baseplate associated genes were found in both genomes, the structure of the baseplate was only observed for phage Bp8p-C virion (Fig. 1C) and not for phiAGATE (16).
Structural proteome analysis of phage Bp8p-C.
The virion proteins of Bp8p-C were separated using SDS-PAGE, and 32 visible bands on the gel were analyzed by MALDI-TOF-TOF MS resulting in the identification of 24 bands (Table 2; see Fig. S2 in the supplemental material). Fourteen proteins were identified from the 24 bands (Table 2), while the other eight bands showed no corresponding protein encoded by the phage genome. Different bands on SDS-PAGE were found to corresponding with the same protein, this might be due to the degradation of phage proteins. Among the bands identified, bands 19, 21, and 22 were annotated as hypothetical proteins (Gp126, Gp62, and Gp138). The encoding genes of Gp126 and Gp138 were found in the genomes of the Bacillus myovirus phages with high similarity, and these two genes were found in phage Bp8p-C genome region that encoded the phage structural protein. The identification of Gp62, Gp126, and Gp138 on Bp8p-C virions suggested that these proteins were phage structural proteins with unknown functions. Bands 7 and 9, with the highest intensity from SDS-PAGE, were identified as the tail sheath protein (Gp122) and the major capsid protein (Gp129), respectively, which are major components of the phage virion with high copy numbers. Gp172, which was annotated as pectin lyase-like protein, was first identified from the phage virions. This protein was the virulence factor of the plant pathogen and took part in the infection of bacteria (45). The pectin lyase-like protein was also reported to facilitate the phage infection (46). The identification of Gp172 on the phage virion made the use of phage Bp8p-C in the control of ginger rhizome rot disease controversial, and the function of Gp172 needs to be further studied. Another protein, Gp173, which was annotated as poly-gamma-glutamate hydrolase, was also first identified as the structural protein of phage. Poly-gamma-glutamate hydrolase can hydrolyze extracellular poly-gamma-glutamate produced by bacteria and facilitate phage infection (49).
TABLE 2.
Characteristics of the Bp8p-C virion-associated protein identified by MALDI-TOF-TOF MSa
| Gene product | Band no. | Molecular mass (kDa) | No. of peptides | Coverage (%) | Predicted function |
|---|---|---|---|---|---|
| Gp62 | 21 | 43.2 | 6 | 17.0 | Hypothetical protein |
| Gp99 | 1, 2, 4, 5 | 121.9 | 48 | 54.3 | Tail protein |
| Gp101 | 23 | 52.3 | 2 | 5.4 | Baseplate protein |
| Gp103 | 24 | 25.9 | 2 | 6.5 | Baseplate protein W |
| Gp108 | 20 | 22.1 | 3 | 6.3 | Minor structural protein 2 |
| Gp111 | 17 | 21.0 | 2 | 15.8 | Tail lysin 1 |
| Gp112 | 3 | 132.0 | 5 | 4.9 | Tail lysin 2 |
| Gp122 | 7, 8, 12-15 | 62.1 | 38 | 64.1 | Tail sheath protein |
| Gp126 | 19 | 31.6 | 3 | 9.3 | Hypothetical protein |
| Gp129 | 9, 10-12, 16, 18, 19 | 52.1 | 20 | 53.0 | Major capsid protein |
| Gp132 | 10 | 62.6 | 2 | 5.4 | Portal protein |
| Gp138 | 22 | 19.9 | 2 | 12.0 | Hypothetical protein |
| Gp172 | 4, 6 | 58.4 | 4.3 | 9.0 | Pectin lyase-like protein |
| Gp173 | 21 | 22.07 | 4 | 33.7 | Poly-gamma-glutamate hydrolase |
For each gene product (Gp), the corresponding band number(s), the molecular mass, the number of peptides identified, the coverage of the identified peptides against the protein sequence, and the putative protein function are listed in Fig. S3 in the supplemental material.
Phage lysogeny.
The plaques formed by phage Bp8p-T exhibited a clear center and a turbid rim (Fig. 1B), suggesting that Bp8p-T could convert to lysogenic state after infection. To evaluate the lysogenic converting ability of phage Bp8p-C and Bp8p-T, strain GR8 was separately infected with the two phages at an MOI of 1.0. After incubation for 4 days, the spores were quantified, and the lysogenic conversion rate was determined by PCR amplification of gene orf27, which revealed that orf27 was present in both phage genomes but not in the genome of GR8 (Fig. 6A). Compared to the control strain GR8, the spore concentrations of strain GR8 infected by the two phages were visibly decreased, and the spore concentration of strain GR8 infected by phage Bp8p-T was higher than that by phage Bp8p-C. PCR amplification of phage gene orf27 showed that the number of positive spores (2.1 × 106 spores/ml) from the Bp8p-T-infected strain was also visibly higher than that from Bp8p-C-infected strain (3.83 × 104 spores/ml) (Fig. 6B). The results suggest that both phages could show latency in GR8, whereas Bp8p-T had higher lysogenic conversion ability than Bp8p-C.
FIG 6.
Assay of lysogeny converting ability of the phage. (A) Detection of orf27 in genomes of strain GR8 and strains infected with phages Bp8p-C or Bp8p-T by PCR amplification. Sterile water was used as control. (B) Determination of lysogeny converting ability of phages Bp8p-C and Bp8p-T. After infection by the phage, the number of spores formed and the calculated number of strains with latent phage were evaluated. The experiment was carried out in triplicate and average values are indicated. (C) Plaques formed by adding the supernatants of GT32, GR8, and GC7. Gradient dilutions of 8-h culture supernatants of GR8, GC7, and GT32 were tested. For each spot, a 1-μl sample was added. GT32 and GC7 indicate phage-infected strain GR8, infected by latent phage Bp8p-T and Bp8p-C, respectively. (D) Growth curves of bacteria. The small graph shows the growth curves from 150 to 300 min by magnifying the vertical axis. GR8 indicated the growth curve of uninfected strain GR8, and T32 and C7 indicated the growth curves of strains GT32 and GC7 with phages Bp8p-T and Bp8p-C latent, respectively.
To determine the stability of the lysogenic state, strain GR8 isolates, GT32 with latent Bp8p-T and GC7 with latent Bp8p-C, were selected. After incubation for 8 h, plaques from the GT32 supernatant were turbid, while that from the GC7 supernatant were clearer. In contrast, no phage was detected from the supernatant of GR8 (Fig. 6C). The plaque morphology from GT32 and GC7 supernatants were similar to that formed by phages Bp8p-T and Bp8p-C, respectively. The growth curves of stains GR8, GT32, and GC7 were assayed (Fig. 6D). The lysogen GT32 and GC7 grew more slowly and weakly than GR8. Compared to GR8, the exponential phase of strain GT32 was delayed about 60 min, but for strain GC7 no typical growth curve was observed, and a slight increase in biomass was observed from 150 to 240 min postinfection. After 240 min, the release of the newly propagated phage Bp8p-C caused the lysis of GC7, while Bp8p-T exhibited higher stability as a lysogenic phage.
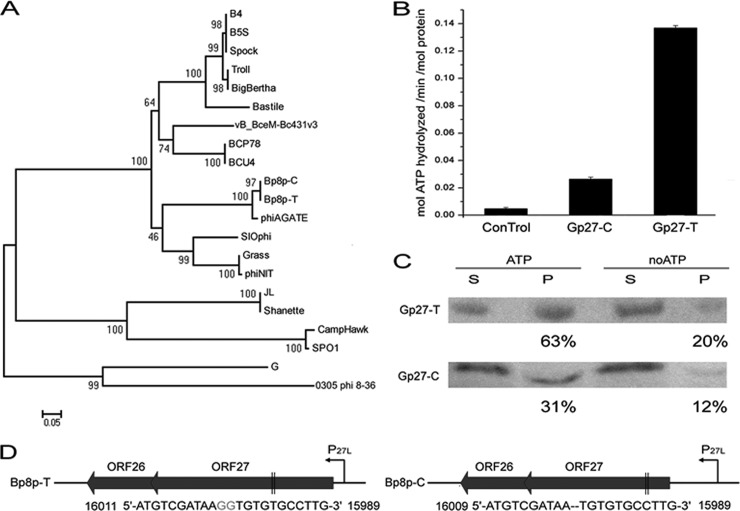
Functional analysis of phage-encoding ParM-like proteins.
Two proteins, Gp27-C and Gp27-T, encoded by the genomes of phage Bp8p-C and Bp8p-T, respectively, were annotated as ParM-like protein, which was similar to the plasmid segregation actin-like motor protein in bacteria. Compared to Gp27-T, two missing guanine nucleotides led to a frameshift mutation of GP27 and encoded a truncated Gp27-C (see Fig. S3 in the supplemental material). Although the genes encoding ParM-like proteins were found in the Bacillus phage genomes with high similarity (Fig. 7A), the exact function of the protein was still unknown. To analyze the function of Gp27-C and Gp27-T, the two proteins were expressed and purified. ATPase activity assay revealed that the Gp27-T had 5-fold-higher ability than Gp27-C to hydrolyze ATP (Fig. 7B), while sedimentation analysis demonstrated that the assembly ability of Gp27-T was also higher than that of Gp27-C (Fig. 7C; Table 1). These results suggest that the truncation of Gp27 resulted in reduction of the ATPase and polymerization abilities, while the truncation of Gp27-C allowed phage Bp8p-C to convert easily from latent to lytic phase and resulted in a change in phage phenotype (Fig. 1A and B). The ParMRC plasmid segregation systems of bacteria requires a DNA-binding protein that tethers the plasmid to the motor protein to mediate partition (5). A small gene orf26 encoding a putative protein containing the helix–turn-helix (HTH) motif that is present in DNA-binding protein (50) was found upstream of orf27 with a 77-bp overlap (Fig. 7D). Based on the location, the small encoding product (110 amino acid residues), and the existence of HTH motif, Gp26 was a possible candidate as a ParR adaptor for Gp27.
FIG 7.
Functional analysis of ParM-like proteins encoded by phage Bp8p-C and Bp8p-T genomes. (A) Phylogenetic tree based on the amino acid sequences of ParM-like proteins from Bacillus phages. (B) Determination of ATPase activity of GP27-C and GP27-T. A final concentration of 0.5 μM protein was used, with buffer serving as a control. (C) Sedimentation analysis of Gp27-C and Gp27-T assembly. After assembling for 30 min and centrifugation, the supernatant (S) and the pellet (P) of the proteins were separated and analyzed by Western blotting. Proteins without ATP (noATP) were used as controls, and the percentages of protein in pellet fractions are indicated. (D) Gene organizations of the putative plasmid segregation associate systems found in Bp8p-C and Bp8p-T genome. The predict promoters (P27L) are indicated with curved arrows, while mutations of orf27 are indicated in gray. The locations of the mutation site in orf27 are indicated between two black vertical lines.
Phage therapy for B. pumilus infection.
Due to the specificity of Bp8p-C and Bp8p-T to infect GR8, they exhibit potential to control ginger rhizome rot disease. To evaluate the ability of the two phages to control ginger rhizome rot disease caused by B. pumilus GR8, the phages were added onto the ginger rhizome slices infected by GR8. The disease severity ratings of the ginger rhizome slices decreased with an increase in phage concentration (Fig. 8). Only mild rot symptom was observed when the concentrations reached 108 PFU/ml for phage Bp8p-C and 109 PFU/ml for Bp8p-T. The control effect by Bp8p-C on rotting of ginger rhizome slices was higher than that by Bp8p-T when the phages were used at the same concentration. Based on the results, phage Bp8p-C might be more effective in the control of ginger rhizome rot disease than phage Bp8p-T.
FIG 8.
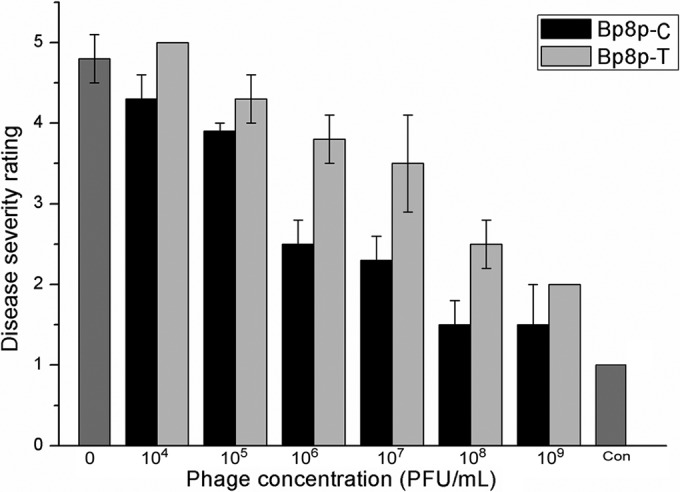
Therapy of ginger rhizome slices rot with Bp8p-C and Bp8p-T. Each ginger rhizome slice was and later infected with 100 μl of log-phase B. pumilus GR8 cell suspension at a concentration of 108 CFU/ml for 5 min and subsequently 100 μl of phage suspension at different concentrations. For the blank control (indicated as Con), 100 μl of sterile water and 100 μl of SM buffer were added.
DISCUSSION
In this study, we characterized the physiology, genome sequences, structural proteome, and lysogenic ability of two phages, Bp8p-C and Bp8p-T, as well as explored the mechanism of actin-like protein affecting phage unstable lysogeny. Although the two phages exhibited different phenotypes in the plaque morphology, there were only three different nucleotides between their genomes. One nucleotide, located upstream of the long terminal repeat region, had no impact on any proteins encoded or regulatory systems. The deletion of two guanine nucleotides in the Bp8p-C genome led to a truncated Gp27, which is a putative plasmid segregation-associated actin-like protein.
The plasmid segregation system is essential for the segregation of some essential plasmids and some plasmids with low copy numbers. In prokaryotes, the actin-like filamentous proteins allow accurate distribution of the plasmids in daughter cells during cell division (51). Actin-like proteins have also been found in several phage genomes such as that of C. botulinum phage c-st (3), B. megaterium phage G (52), Pseudomonas chlororaphis phage 201ϕ2-1 (6), and B. pumilus phage phiAGATE (16). The actin-like protein encoded by c-st genome belongs to the type III plasmid segregation system and has been regarded as one of the factors causing unstable lysogeny. However, no experimental data have been reported about the role of the protein in regulating lysogeny in phage c-st. Based on the statistical relationship between the temperate feature and the existence of the parA and parB genes, one study on mycobacteriophages has also led to the speculation that the existence of plasmid segregation-associated protein was responsible for phage lysogeny (53). In that study, the difference between the putative plasmid segregation-associated actin-like proteins, Gp27-C and Gp27-T, encoded by phage Bp8p-C and Bp8p-T, respectively, led to different phenotypes of phage lysogeny. Based on previous reports and the results of the present study, the actin-like protein encoded by phage can regulate the life cycle of the phage. We speculated that the putative plasmid segregation system encoded by phage genome was helpful in leading the phage genomes to distribute into daughter cells as phage-plasmids. This strategy for phage genome will interdict the process of phage particle synthesis and finally lead to “pseudolysogeny” (3). The truncation of the putative actin-like plasmid segregation-associated protein encoded by Bp8p-C genome caused a reduction in protein function, and the phage genomes were packed into the phage capsid with a higher frequency. The experimental data demonstrate a direct relationship between the actin-like protein and the phage lysogenic ability. The finding that the truncated putative actin-like plasmid segregation-associated protein affected the phage lysogenic ability revealed a conceivable novel unstable lysogenic mechanism for the phage.
Lytic phages propagate by infecting bacteria and causing lysis of the host cells, whereas lysogenic phages can remain latent within the host cells (1). The constituents of phage, mainly composed of proteins and nucleic acids, result in its instability (54). As the viral predator of bacteria, species conservation of the host strain is essential for phage survival. By converting to the lysogenic state, the phage can achieve species conservation by replicating as part of the host chromosome or as individual plasmid. One important characteristic of Bacillus strains is the formation of spores for survival during adverse growth conditions. Because of its ability to convert into latent phage, the Bacillus phage can achieve species conservation by incorporating itself into the host spore. The putative plasmid segregation protein is widely found in the genome of Bacillus myovirus (16, 55) and might be a common survival mechanism of the Bacillus myovirus. A further functional analysis of the putative phage-encoding plasmid segregation protein in vivo will illustrate how the protein regulates the state of phage genome, whether the phage genomes are packaged into phage capsids or distribute into daughter cells as phage-plasmid during cell division.
Phages encode certain proteins to facilitate infection. In the present study, two proteins were identified in the virion proteome of Bp8p-C that could possibly have similar functions. One was a poly-gamma-glutamate hydrolase, which has been reported to cause hydrolysis of the extracellular poly-gamma-glutamate produced by the bacterium (49), and the other was a pectin lyase-like protein, which can disrupt the bacterial biofilm by degrading the matrix polysaccharides (46). Despite the function of pectin lyase-like protein as virulence factor of plant-pathogenic bacteria, both proteins may facilitate the infection of the phage. Similar proteins are also found to be encoded by several other phages (16), suggesting that these two genes are extensively found in the genome of Bacillus phages, and this might be an adaptive mechanism for the Bacillus phages to infect their host strains.
The annotation of ORFs with unknown function is one of the major challenges of phage molecular biology. Three proteins (Gp62, Gp126, and Gp138) with unknown functions were found to be the components of phage virion by proteomic analysis, suggesting that these three proteins were potential phage structural proteins. Although the exact function of the three proteins are still unclear, our proteomic analysis here has provided useful information for their functional annotation.
Due to the ability to lyse plant-pathogenic bacteria with high specificity, several phages are regarded as promising agents for plant disease control. Phages Bp8p-C and Bp8p-T exhibited high activity in controlling ginger rhizome rot disease. Compared to other phages used for bacterial pathogen control (56), Bp8p-C showed a larger burst size, which would benefit its application in ginger rhizome rot disease control. However, the identification of a pectin lyase-like protein on the phage virions made the use of these two phages controversial. With respect to the bacterium, a pectin lyase-like protein-encoding gene carried by the phage might enhance the fitness of the host strain by serving as a bacterial virulence factor to increase the infection ability of the host strain (57), but from the vantage point of the phage, pectin lyase-like protein might enhance the infection by the phage. Studies on the function of pectin lyase-like protein will illustrate whether these two phages are suitable for direct application without further manipulation or a genetic modification of the phage will help overcome this challenge.
Supplementary Material
ACKNOWLEDGMENTS
We thank Lei Zhang from the Roche 454 Genome Sequence Platform of State Key Laboratory of Virology, Wuhan Institute of Virology, Chinese Academy of Sciences, for help in phage genome sequence.
This study was supported by the projects of the Chinese Academy of Sciences (KSZD-EW-Z-021-2-2) and the National Natural Science Foundation of China (no. 31170123).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02889-14.
REFERENCES
- 1.Kim M, Ryu S. 2013. Antirepression system associated with the life cycle switch in the temperate Podoviridae phage SPC32H. J Virol 87:11775–11786. doi: 10.1128/JVI.02173-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reichardt LF. 1975. Control of bacteriophage lambda-repressor synthesis: regulation of maintenance pathway by Cro and Ci products. J Mol Biol 93:289–309. doi: 10.1016/0022-2836(75)90133-3. [DOI] [PubMed] [Google Scholar]
- 3.Sakaguchi Y, Hayashi T, Kurokawa K, Nakayama K, Oshima K, Fujinaga Y, Ohnishi M, Ohtsubo E, Hattori M, Oguma K. 2005. The genome sequence of Clostridium botulinum type C neurotoxin-converting phage and the molecular mechanisms of unstable lysogeny. Proc Natl Acad Sci U S A 102:17472–17477. doi: 10.1073/pnas.0505503102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oliva MA, Martin-Galiano AJ, Sakaguchi Y, Andreu JM. 2012. Tubulin homolog TubZ in a phage-encoded partition system. Proc Natl Acad Sci U S A 109:7711–7716. doi: 10.1073/pnas.1121546109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Salje J, Gayathri P, Lowe J. 2010. The ParMRC system: molecular mechanisms of plasmid segregation by actin-like filaments. Nat Rev Microbiol 8:683–692. doi: 10.1038/nrmicro2425. [DOI] [PubMed] [Google Scholar]
- 6.Kraemer JA, Erb ML, Waddling CA, Montabana EA, Zehr EA, Wang HN, Nguyen K, Pham DSL, Agard DA, Pogliano J. 2012. A phage tubulin assembles dynamic filaments by an atypical mechanism to center viral DNA within the host cell. Cell 149:1488–1499. doi: 10.1016/j.cell.2012.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schumacher MA, Glover TC, Brzoska AJ, Jensen SO, Dunham TD, Skurray RA, Firth N. 2007. Segrosome structure revealed by a complex of ParR with centromere DNA. Nature 450:1268-U1215. doi: 10.1038/nature06392. [DOI] [PubMed] [Google Scholar]
- 8.Fritze D. 2004. Taxonomy of the genus Bacillus and related genera: the aerobic endospore-forming bacteria. Phytopathology 94:1245–1248. doi: 10.1094/PHYTO.2004.94.11.1245. [DOI] [PubMed] [Google Scholar]
- 9.Myresiotis CK, Karaoglanidis GS, Vryzas Z, Papadopoulou-Mourkidou E. 2012. Evaluation of plant-growth-promoting rhizobacteria, acibenzolar-S-methyl and hymexazol for integrated control of Fusarium crown and root rot on tomato. Pest Manag Sci 68:404–411. doi: 10.1002/ps.2277. [DOI] [PubMed] [Google Scholar]
- 10.Logan NA. 2012. Bacillus and relatives in food-borne illness. J Appl Microbiol 112:417–429. doi: 10.1111/j.1365-2672.2011.05204.x. [DOI] [PubMed] [Google Scholar]
- 11.Kimouli M, Vrioni G, Papadopoulou M, Koumaki V, Petropoulou D, Gounaris A, Friedrich AW, Tsakris A. 2012. Two cases of severe sepsis caused by Bacillus pumilus in neonatal infants. J Med Microbiol 61:596–599. doi: 10.1099/jmm.0.033175-0. [DOI] [PubMed] [Google Scholar]
- 12.Galal AA, El-Bana AA, Janse J. 2006. Bacillus pumilus, a new pathogen on mango plants. Egypt J Phytopathol 34:17–29. [Google Scholar]
- 13.Li B, Qiu W, Tan Q, Su T, Fang Y, Xie G. 2009. Association of a Bacillus species with leaf and twig dieback of Asian pear (Pyrus pyrifolia) in China. J Plant Pathol 91:705–708. [Google Scholar]
- 14.Peng Q, Yuan Y, Gao M. 2013. Bacillus pumilus, a novel ginger rhizome rot pathogen in China. Plant Dis 97:1308–1315. doi: 10.1094/PDIS-12-12-1178-RE. [DOI] [PubMed] [Google Scholar]
- 15.Bramucci MG, Keggins KM, Lovett PS. 1977. Bacteriophage conversion of spore-negative mutants to spore-positive in Bacillus pumilus. J Virol 22:194–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barylski J, Nowicki G, Gozdzicka-Jozefiak A. 2014. The discovery of ϕAGATE, a novel phage infecting Bacillus pumilus, leads to new insights into the phylogeny of the subfamily Spounavirinae. PLoS One 9:e86632. doi: 10.1371/journal.pone.0086632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lorenz L, Lins B, Barrett J, Montgomery A, Trapani S, Schindler A, Christie GE, Cresawn SG, Temple L. 2013. Genomic characterization of six novel Bacillus pumilus bacteriophages. Virology 444:374–383. doi: 10.1016/j.virol.2013.07.004. [DOI] [PubMed] [Google Scholar]
- 18.Mageeney C, Pope WH, Harrison M, Moran D, Cross T, Jacobs-Sera D, Hendrix RW, Dunbar D, Hatfull GF. 2012. Mycobacteriophage Marvin: a new singleton phage with an unusual genome organization. J Virol 86:4762–4775. doi: 10.1128/JVI.00075-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yuan YH, Gao MY, Wu DD, Liu PM, Wu Y. 2012. Genome characteristics of a novel phage from Bacillus thuringiensis showing high similarity with phage from Bacillus cereus. PLoS One 7:e37557. doi: 10.1371/journal.pone.0037557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thomas JA, Hardies SC, Rolando M, Hayes SJ, Lieman K, Carroll CA, Weintraub ST, Serwer P. 2007. Complete genomic sequence and mass spectrometric analysis of highly diverse, atypical Bacillus thuringiensis phage 0305 phi 8-36. Virology 368:405–421. doi: 10.1016/j.virol.2007.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lynch KH, Stothard P, Dennis JJ. 2010. Genomic analysis and relatedness of P2-like phages of the Burkholderia cepacia complex. BMC Genomics 11:599. doi: 10.1186/1471-2164-11-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Catalao MJ, Gil F, Moniz-Pereira J, Pimentel M. 2010. The mycobacteriophage Ms6 encodes a chaperone-like protein involved in the endolysin delivery to the peptidoglycan. Mol Microbiol 77:672–686. doi: 10.1111/j.1365-2958.2010.07239.x. [DOI] [PubMed] [Google Scholar]
- 23.Garcia P, Martinez B, Obeso JM, Lavigne R, Lurz R, Rodriguez A. 2009. Functional genomic analysis of two Staphylococcus aureus phages isolated from the dairy environment. Appl Environ Microbiol 75:7663–7673. doi: 10.1128/AEM.01864-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marchler-Bauer A, Zheng CJ, Chitsaz F, Derbyshire MK, Geer LY, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Lanczycki CJ, Lu F, Lu SN, Marchler GH, Song JS, Thanki N, Yamashita RA, Zhang DC, Bryant SH. 2013. CDD: conserved domains and protein three-dimensional structure. Nucleic Acids Res 41:D348–D352. doi: 10.1093/nar/gks1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, Pang N, Forslund K, Ceric G, Clements J, Heger A, Holm L, Sonnhammer ELL, Eddy SR, Bateman A, Finn RD. 2012. The Pfam protein families database. Nucleic Acids Res 40:D290–D301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Soding J. 2005. Protein homology detection by HMM-HMM comparison. Bioinformatics 21:951–960. doi: 10.1093/bioinformatics/bti125. [DOI] [PubMed] [Google Scholar]
- 27.Stothard P, Wishart DS. 2005. Circular genome visualization and exploration using CGView. Bioinformatics 21:537–539. doi: 10.1093/bioinformatics/bti054. [DOI] [PubMed] [Google Scholar]
- 28.Tatusov RL, Koonin EV, Lipman DJ. 1997. A genomic perspective on protein families. Science 278:631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- 29.Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res 33:W686–W689. doi: 10.1093/nar/gki366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Benson G. 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res 27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Siguier P, Perochon J, Lestrade L, Mahillon J, Chandler M. 2006. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res 34:D32–D36. doi: 10.1093/nar/gkj014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krumsiek J, Arnold R, Rattei T. 2007. Gepard: a rapid and sensitive tool for creating dotplots on genome scale. Bioinformatics 23:1026–1028. doi: 10.1093/bioinformatics/btm039. [DOI] [PubMed] [Google Scholar]
- 33.Darling ACE, Mau B, Blattner FR, Perna NT. 2004. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res 14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zafar N, Mazumder R, Seto D. 2002. CoreGenes: a computational tool for identifying and cataloging “core” genes in a set of small genomes. BMC Bioinformatics 3:12. doi: 10.1186/1471-2105-3-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum-parsimony methods. Mol Biol. Evol 28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gao MY, Li RS, Dai SY, Wu Y, Yi D. 2008. Diversity of Bacillus thuringiensis strains from soil in China and their pesticidal activities. Biol Control 44:380–388. doi: 10.1016/j.biocontrol.2007.11.011. [DOI] [Google Scholar]
- 37.Sambrook JFE, Maniatis T. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 38.Popp D, Xu WJ, Narita A, Brzoska AJ, Skurray RA, Firth N, Goshdastider U, Maeda Y, Robinson RC, Schumacher MA. 2010. Structure and filament dynamics of the pSK41 actin-like ParM protein implications for plasmid DNA segregation. J Biol Chem 285:10130–10140. doi: 10.1074/jbc.M109.071613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hara F, Yamashiro K, Nemoto N, Ohta Y, Yokobori SI, Yasunaga T, Hisanaga SI, Yamagishi A. 2007. An actin homolog of the archaeon Thermoplasma acidophilum that retains the ancient characteristics of eukaryotic actin. J Bacteriol 189:2039–2045. doi: 10.1128/JB.01454-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lindner P, Bauer K, Kremmer E, Krebber C, Honegger A, Klinger B, Mocikat R, Pluckthun A. 1997. Specific detection of his-tagged proteins with recombinant anti-His tag scFv-phosphatase or scFv-phage fusions. Biotechniques 22:140–149. [DOI] [PubMed] [Google Scholar]
- 41.Moak M, Molineux IJ. 2004. Peptidoglycan hydrolytic activities associated with bacteriophage virions. Mol Microbiol 51:1169–1183. doi: 10.1046/j.1365-2958.2003.03894.x. [DOI] [PubMed] [Google Scholar]
- 42.Lavigne R, Noben JP, Hertveldt K, Ceyssens PJ, Briers Y, Dumont D, Roucourt B, Krylov VN, Mesyanzhinov VV, Robben J, Volckaert G. 2006. The structural proteome of Pseudomonas aeruginosa bacteriophage phi KMV. Microbiology 152:529–534. doi: 10.1099/mic.0.28431-0. [DOI] [PubMed] [Google Scholar]
- 43.Siew N, Azaria Y, Fischer D. 2004. The ORFanage: an ORFan database. Nucleic Acids Res 32:D281–D283. doi: 10.1093/nar/gkh116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Handtke S, Schroeter R, Jurgen B, Methling K, Schluter R, Albrecht D, van Hijum SAFT, Bongaerts J, Maurer KH, Lalk M, Schweder T, Hecker M, Voigt B. 2014. Bacillus pumilus reveals a remarkably high resistance to hydrogen peroxide provoked oxidative stress. PLoS One 9:e85625. doi: 10.1371/journal.pone.0085625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lombard V, Bernard T, Rancurel C, Brumer H, Coutinho PM, Henrissat B. 2010. A hierarchical classification of polysaccharide lyases for glycogenomics. Biochem J 432:437–444. doi: 10.1042/BJ20101185. [DOI] [PubMed] [Google Scholar]
- 46.Thompson JE, Pourhossein M, Waterhouse A, Hudson T, Goldrick M, Derrick JP, Roberts IS. 2010. The K5 lyase KflA combines a viral tail spike structure with a bacterial polysaccharide lyase mechanism. J Biol Chem 285:23963–23969. doi: 10.1074/jbc.M110.127571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Catalao MJ, Gil F, Moniz-Pereira J, Sao-Jose C, Pimentel M. 2013. Diversity in bacterial lysis systems: bacteriophages show the way. FEMS Microbiol Rev 37:554–571. doi: 10.1111/1574-6976.12006. [DOI] [PubMed] [Google Scholar]
- 48.Vandersteegen K, Kropinski AM, Nash JHE, Noben JP, Hermans K, Lavigne R. 2013. Romulus and Remus, two phage isolates representing a distinct clade within the Twortlikevirus genus, display suitable properties for phage therapy applications. J Virol 87:3237–3247. doi: 10.1128/JVI.02763-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fujimoto Z, Kimura K. 2012. Crystal structure of bacteriophage Phi NIT1 zinc peptidase PghP that hydrolyzes gamma-glutamyl linkage of bacterial poly-gamma-glutamate. Proteins 80:722–732. doi: 10.1002/prot.23229. [DOI] [PubMed] [Google Scholar]
- 50.Aravind L, Anantharaman V, Balaji S, Babu MM, Iyer LM. 2005. The many faces of the helix-turn-helix domain: transcription regulation and beyond. FEMS Microbiol Rev 29:231–262. doi: 10.1016/j.femsre.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 51.Ebersbach G, Gerdes K. 2005. Plasmid segregation mechanisms. Annu Rev Genet 39:453–479. doi: 10.1146/annurev.genet.38.072902.091252. [DOI] [PubMed] [Google Scholar]
- 52.Kristensen DM, Cai XX, Mushegian A. 2011. Evolutionarily conserved orthologous families in phages are relatively rare in their prokaryotic hosts. J Bacteriol 193:1806–1814. doi: 10.1128/JB.01311-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stella EJ, Franceschelli JJ, Tasselli SE, Morbidoni HR. 2013. Analysis of novel mycobacteriophages indicates the existence of different strategies for phage inheritance in mycobacteria. PLoS One 8:e56384. doi: 10.1371/journal.pone.0056384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mizuno CM, Rodriguez-Valera F, Kimes NE, Ghai R. 2013. Expanding the marine virosphere using metagenomics. PLoS Genet. 9:e1003987. doi: 10.1371/journal.pgen.1003987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee JH, Shin H, Son B, Ryu S. 2012. Complete genome sequence of Bacillus cereus bacteriophage BCP78. J Virol 86:637–638. doi: 10.1128/JVI.06520-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bandara N, Jo J, Ryu S, Kim KP. 2012. Bacteriophages BCP1-1 and BCP8-2 require divalent cations for efficient control of Bacillus cereus in fermented foods. Food Microbiol 31:9–16. doi: 10.1016/j.fm.2012.02.003. [DOI] [PubMed] [Google Scholar]
- 57.Moran NA, Degnan PH, Santos SR, Dunbar HE, Ochman H. 2005. The players in a mutualistic symbiosis: insects, bacteria, viruses, and virulence genes. Proc Natl Acad Sci U S A 102:16919–16926. doi: 10.1073/pnas.0507029102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.