Figure 1.
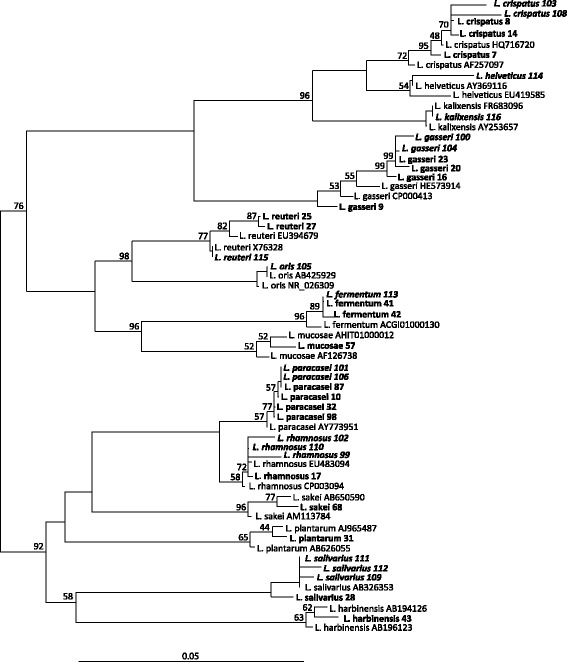
Phylogenetic tree based on the analysis of 16S rRNA gene sequences derived from oral and vaginal isolates. 16S rDNA sequences were aligned using the SINA plugin (ARB software package) and distances were calculated using the neighbor-joining method with Jukes-Cantor correction. Reference sequences with accession numbers are in regular type, sample sequences of vaginal isolates in bold type and of oral isolates in bold/italics type. Sample strains are named according to their identification by 16S rDNA sequencing results. Bootstrap values (from 1000 replicates) greater than 40% are shown at the branch points. The bar indicates 5% sequence divergence.
