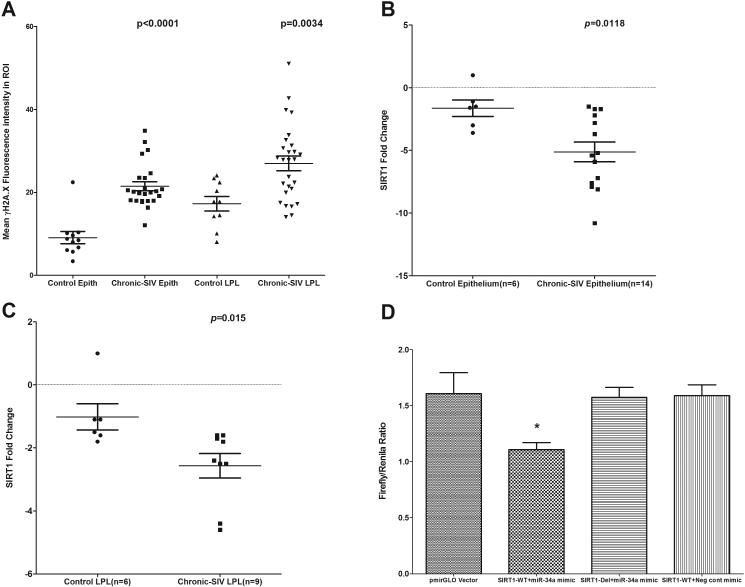
FIGURE 4.
Quantitation of cells and regions of interest (ROI) labeled by phospho-γH2A.X. was performed using Volocity 5.5 software after capturing images on a Leica confocal microscope. Several ROI were hand drawn on the epithelial and LPL regions in the images from colon of chronic SIV-infected and uninfected macaques (A). Data were analyzed using non-parametric Wilcoxon’s rank sum test. qRT-PCR revealed significantly decreased expression of SIRT1 mRNA in both colonic epithelium (p=0.0118) (B) and LPL (p=0.015) (C) compartments of chronic SIV-infected compared to uninfected control macaques. Data were analyzed using the non-parametric Wilcoxon’s rank sum test. The error bars represent standard error of mean fold change within each group. miR-34a physically associates with the 3’ UTR of rhesus macaque SIRT1 mRNA (D). The SIRT1 wild type and deleted 3′UTR sequences were inserted into the multiple cloning sites situated in the 3′ end of firefly luciferase gene in the pmirGLO vector. HEK293 cells were co-transfected with 100 nM miR-34a or negative control mimic and 100 ng of luciferase reporter constructs containing wild type (WT) or deleted (Del) SIRT1 3′UTR sequences. Firefly and Renila luciferase activities were detected using the Dual-Glo luciferase assay system 72 h after transfection. The ratio of luciferase activities (Firefly/Renilla) was calculated and normalized to the wells transfected with only unmanipulated pmir-GLO vector. Single asterisk (*) indicates statistical significance (p< 0.05) compared to cells transfected with unmanipulated pmirGLO vector.