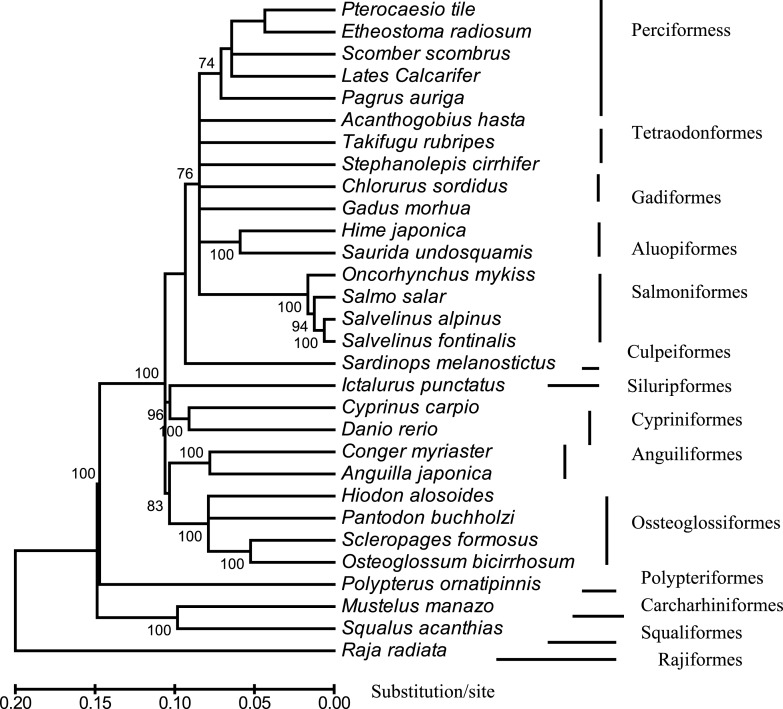
Fig. 3.
An NJ-tree reconstructed with MEGA, showing the evolutionary position of Lates calcrifer relative to that of 30 other fish species belonging to 14 suborders. The tree is based on concatenated amino acids sequences of 12 protein-coding genes. The scale bar is shown under the tree, whereas the bootstrap value (>50%) of 1,000 replicates is listed. The analyses included the following 30 species: Pterocaesio tile (NC–004408), Pagrus auriga (NC–005146), Etheostoma radiosum (NC–005254), Scomber, scombrus (NC–006398), Chlorurus sordidus (NC–006355), Acanthogobius hasta (NC–006131), Takifugu rubripes (AJ421455), Stephanolepis cirrhifer (NC–003177), Gadus morhua (X99772), Hime japonica (AB047821), Saurida undosquamis (NC–003162), Oncorhynchus mykiss (L29771), Salmo salar (U12143), Salvelinus alpinus (AF154851), Salvelinus fontinalis (AF154850), Sardinops melanostictus (AB032554), Ictalurus punctatus (AF482987), Cyprinus carpio (X61010), Danio rerio (AC024175), Conger myriaster (AB038381), Anguilla japonica (AB038556), Hiodon alosoides (AP004356), Pantodon buchholzi (AB043068), Scleropages formosus (DQ023143), Osteoglossum bicirrhosum (AB043025), Mustelus manazo (AB015962), Squalus acanthias (Y18134), Raja radiate (AF106038), Polypterus ornatipinnis (U62532) and Lates calcarifer (DQ010541).