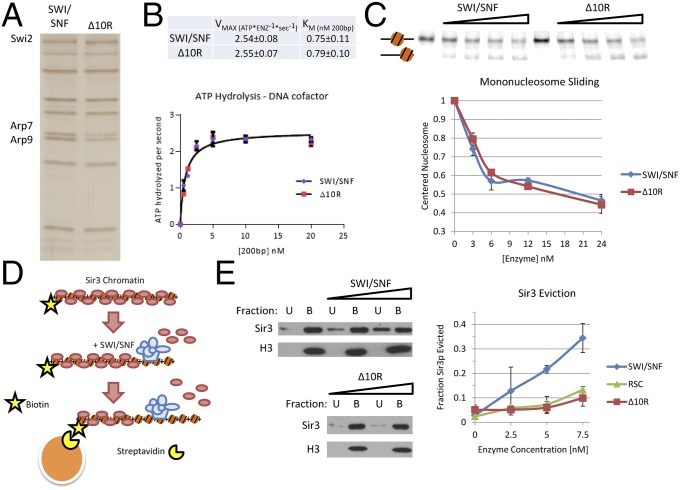
Fig. 4.
Swi2p–Sir3p contacts are required for eviction of Sir3p from nucleosomes. (A) SDS/PAGE analysis of SWI/SNF and SWI/SNF–Δ10R complexes, visualized by silver staining. Equal levels of ATPase activity were loaded for each enzyme. (B) DNA-stimulated ATPase kinetics of SWI/SNF and SWI/SNF–Δ10R are equivalent. ATPase reactions were performed with varying concentrations of DNA cofactor, and hydrolysis rates were fit to Michaelis–Menten kinetic parameters. (C) Mononucleosome mobilization by SWI/SNF and SWI/SNF–Δ10R enzymes is equivalent. Varying concentrations of enzymes were incubated with a mononucleosome positioned in the center of a radiolabeled, 282-bp DNA fragment harboring a 601 positioning sequence. Predicted positions of mononucleosomes are indicated to the left. (Upper) Gel. (Lower) Quantification (error bars reflect SD). (D) Schematic of the chromatin capture assay. Biotinylated nucleosomal arrays are bound to Sir3p, incubated with chromatin-remodeling enzyme and ATP and captured on streptavidin-coated magnetic beads. Chromatin-bound B and unbound U are assayed by Western blotting. (E) SWI/SNF–Δ10R is defective for Sir3p eviction from nucleosomes. Increasing amounts of chromatin-remodeling enzyme were incubated with Sir3p-bound nucleosomal array, and Sir3p eviction into the chromatin-unbound fraction U was measured by Western blotting. (Left) Representative blots. (Right) Quantification.