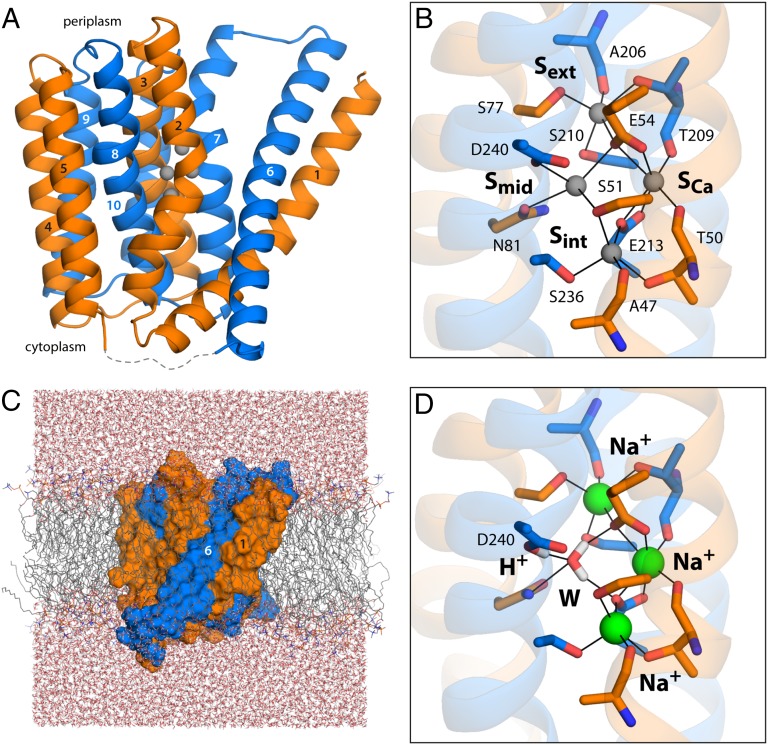
Fig. 1.
Structure of the Na+/Ca2+ exchanger from M. jannaschii in an outward-facing conformation. (A) The architecture of NCX_Mj (PDB entry 3V5U) consists of two repeats of five transmembrane helices each (orange, marine cartoons), which adopt opposite orientations in the membrane (the cytoplasmic side is below). Four putative ion-binding sites (indicated by gray spheres) are found halfway across the transmembrane region, flanked by helices TM2–TM3 and TM7–TM8. The structures of the five-helix repeats are highly similar but not identical (Fig. S1). The major differences are the orientation of TM1/TM6 relative to the rest of the repeat, and a bend in the periplasmic half of TM7, not observed in the equivalent region of TM2. These structural asymmetries likely underlie the ability of the exchanger to alternatively adopt outward- and inward-facing conformations (24). (B) Close-up of the four putative ion-binding sites. The side-chain and backbone groups lining these sites are highlighted; the hypothetical ion coordination contacts are indicated in each case. (C) All-atom simulation model of NCX_Mj embedded in a phospholipid membrane. The membrane consists of 208 POPC molecules, and protein and membrane are hydrated by ∼14,900 water molecules. Chloride ions (omitted for clarity) were also included to counter the net charge of the ion–protein complex. The overall system size is ∼77,300 atoms. (D) Ion configuration that is most likely to correspond to the crystal structure of NCX_Mj, according to the structural and thermodynamic analyses described in Figs. 2 and 3. A water molecule (W) occupies the Smid site, while Na+ ions occupy Sext, SCa, and Sint. Note that D240 is protonated, and that the side-chain carboxamide group of N81 is inverted relative to the original assignment. The figure shows the conformation that is most frequently observed in a 200-ns MD simulation, according to an RMSD-based clustering analysis comprising the residues highlighted in B.