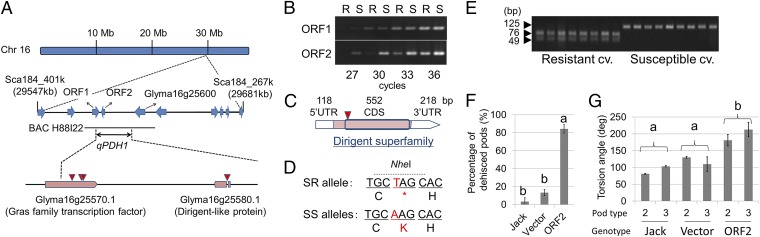
Fig. 2.
Map-based cloning of cultivated soybean [Glycine max Merr. (L.)] qPDH1. (A) Predicted ORFs in the previously determined candidate region of chromosome 16 (blue arrows) and predicted ORFs in the region delimited in the present study (pink boxes). BAC H88I22 indicates a BAC clone carrying the qPDH1 locus of a shattering-susceptible (SS) cultivar, Misuzudaizu. Red triangles indicate positions of single nucleotide polymorphisms (SNPs) between Hayahikari [shattering-resistant (SR)] and Toyomusume (SS). (B) Results from semiquantitative RT-PCR for transcripts of ORF1 and ORF2 in pod walls of near-isogenic lines for qPDH1. R and S indicate lines 85R and 85S, respectively. (C) cDNA structure of ORF2 of Toyomusume. (D) DNA and deduced amino acid sequences around the SNP. Red letters indicate the SNP and the resulting amino acid residue or termination signal. The SNP can be recognized by a restriction enzyme, Nhe I. (E) PCR–RFLP genotyping of SR and SS cultivars at the SNP using Nhe I. SR cultivars (leftmost lanes) and SS cultivars (rightmost lanes) are listed in SI Materials and Methods. In these cultivars, the presence of an SR or SS allele at qPDH1 on chromosome 16 was suggested in this study or previous studies (14, 15, 17, 48, 56). (F) Percentages of pod dehiscence of an SR cultivar, Jack, plants, which were nontransformed, transformed only with the vector, or transformed with ORF2 from Toyomusume, at 30% RH (mean ± SE; n = 3, 2, and 8). Different letters indicate significant differences (P < 0.001). Although nontransformed Jack plants were grown in a different growth chamber, pod dehiscence was simultaneously monitored in the same chamber. (G) Torsion angles of dehisced pod walls of two-seeded (bar 2) or three-seeded (bar 3) pods of nontransformed Jack plants, Jack plants transformed only with the vector, or Jack plants transformed with ORF2 from Toyomusume at 30% RH (mean ± SE; n = 3, 3, 2, 2, 8, and 6). Different letters indicate significant genotypic differences (P < 0.05), detected by two-way ANOVA.