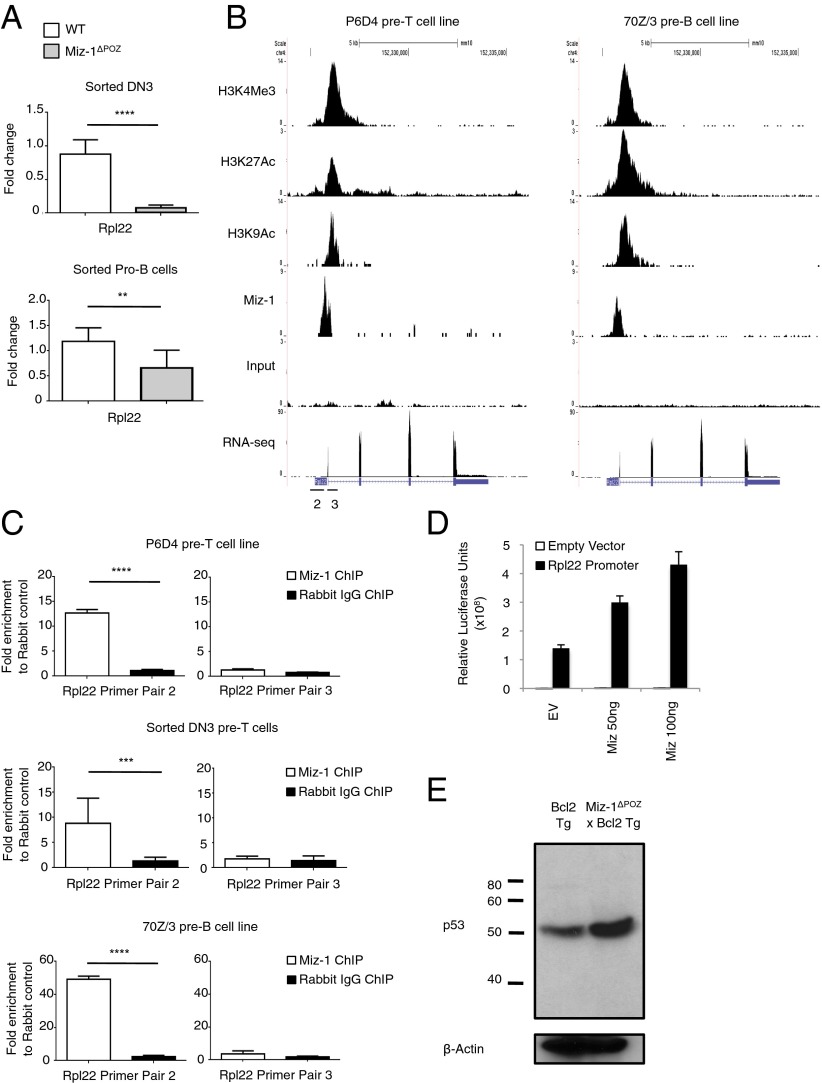
Fig. 6.
Miz-1 regulates the expression of Rpl22 in DN3 pre-T cells and pro-B cells. (A) Rpl22 mRNA expression was assessed in sorted DN3 pre-T cells (Upper) and sorted pro-B cells (Lower) from WT and Miz-1ΔPOZ mice. Data are averaged from three independent experiments and are presented as mean ± SD. (B) ChIP-seq experiments for Miz-1 and histone activation marks from P6D4 murine pre-T cells (Right) and 70Z/3 murine pre-B cells (Left). Shown is the Rpl22 locus. Scale is in number of RPM. Primer pairs were designed in the promoter (2) or first exon (3) of Rpl22 to determine Miz-1 binding by ChIP. (C) ChIP-qPCR experiments to determine binding of Miz-1 to the promoter of Rpl22 in P6D4 murine pre-T cells (Top), sorted WT DN3 pre-T cells (Middle), and 70Z/3 murine pre-B cells (Bottom). Graphs show fold enrichment of anti–Miz-1 ChIP over rabbit IgG control ChIP. Data represent as average fold change ± SD from at least three independent experiments. (D) 293T cells were transfected with the human Rpl22 promoter fused to luciferase and pcDNA3.1 empty vector (EV) or pcDNA3.1 with human Miz-1 in varying concentrations. Data were normalized for transfection using β-galactosidase. Data are presented as average relative luciferase units ± SD and are representative of three independent experiments. (E) Whole protein extracts from total thymus of Bcl2 Tg or Miz-1ΔPOZ × Bcl2 Tg mice were evaluated for p53 expression by Western blot analysis. Data are representative of at least three independent experiments.