FIGURE 1.
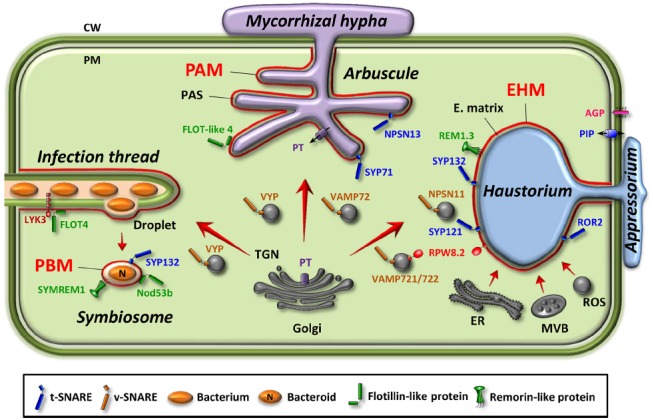
Perimicrobial membranes in three types of plant–microbe interactions. Location of proteins specifically targeted to or excluded from the perimicrobial membranes are indicated. Endomembrane compartments or vesicles involved are illustrated. Small GTPases and exocyst subunits are not represented. AGP, arabinogalactan protein; CW, cell wall; EHM, extrahaustorial membrane; E. matrix, extrahaustorial matrix; ER, endoplasmic reticulum; FLOT, flotillin; LYK3, lysM-family receptor-like kinase from Medicago; MVB, multivesicular body; N, fixed-nitrogen; Nod53b, nodule-specific 53-kDa protein from soybean; NPSN, novel plant SNARE, PAM, periarbuscular membrane; PAS, periarbuscular space; PBM, peribacteroid membrane; PIP, plasma membrane intrinsic protein (aquaporin); PM, plasma membrane; PT, phosphate transporter; REM, remorin; ROR2, required for mlo-specified resistance2 syntaxin; ROS, reactive oxygen species; RPW8.2, resistance powdery mildew8.2; SYMREM, symbiotic remorin1; SYP, syntaxin of plant; TGN, trans-Golgi network; VAMP, vesicle-associated membrane protein; VYP, vapyrin.
