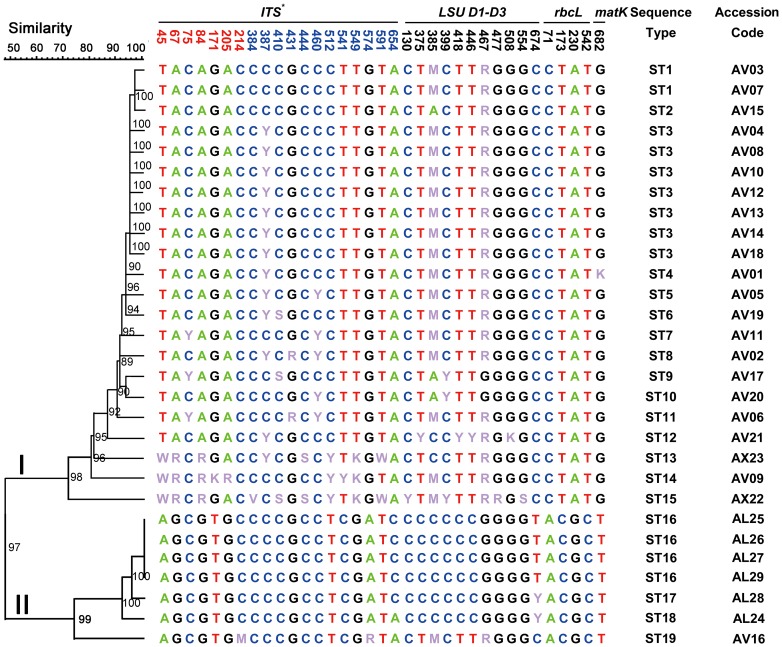
Figure 2. Phylogenetic tree of 29 Amomi Fructus landraces constructed by the UPGMA method based on 35 single nucleotide polymorphisms (SNPs) from four loci (ITS, LSU D1–D3, rbcL, and matK). Numbers on the UPGMA tree branches are bootstrap values (1,000 replicates).
Numbers above the bases indicate the position of SNPs in each locus. *The SNPs of ITS included ITS1 (red numbers) and ITS2 (blue numbers). Heterozygous sites were defined according to IUPAC, i.e., W = A/T; M = A/C; R = A/G; Y = C/T; S = G/C.