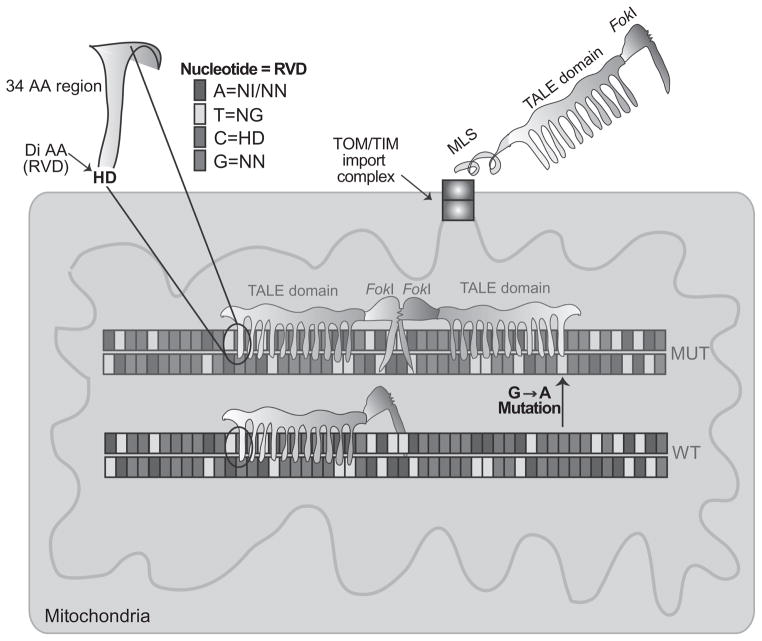
Figure 18.4.
Development of mito-TALENs to alter mtDNA heteroplasmy in vivo. TALENs are enzymes used by the Xanthomonas bacteria to bind and modify specific DNA regions. A nonspecific FokI cleavage domain can be fused to the TALE DNA-binding domain to create a nuclease. However, FokI requires dimerization, and therefore, two TALEN monomers are required for effective cleavage. This requirement provides increased specificity for the DSB. The sequence that binds DNA corresponds to an approximate 34-amino acid sequence: LTPEQVVAIASHDGGKQALETVQRLLPVLC QAHG. The residues at the 12th and the 13th positions are known as repeat variable di-residue or RVD. The figure illustrates how different RVDs are specific to one of the four bases. To direct TALENs to mitochondria, we fused a mitochondrial localization signal (MLS) at the N-terminus of the fusion protein. The TALENs are imported through the TOM/TIM import complexes unfolded, but gain their final folding in the mitochondrial matrix, with the help of molecular chaperones. The MLS is cleaved by a mitochondrial peptidase and the mature protein is ready to bind DNA. When dimers of FokI are formed, the mito-TALEN cleaves the mtDNA haplotype, which is mostly degraded. To target mutant mtDNAs that differ by a single-base pair (in this example, a G>A transition), one of the TALEN monomers must be able to bind only to the mutated sequence and not to the wild type.