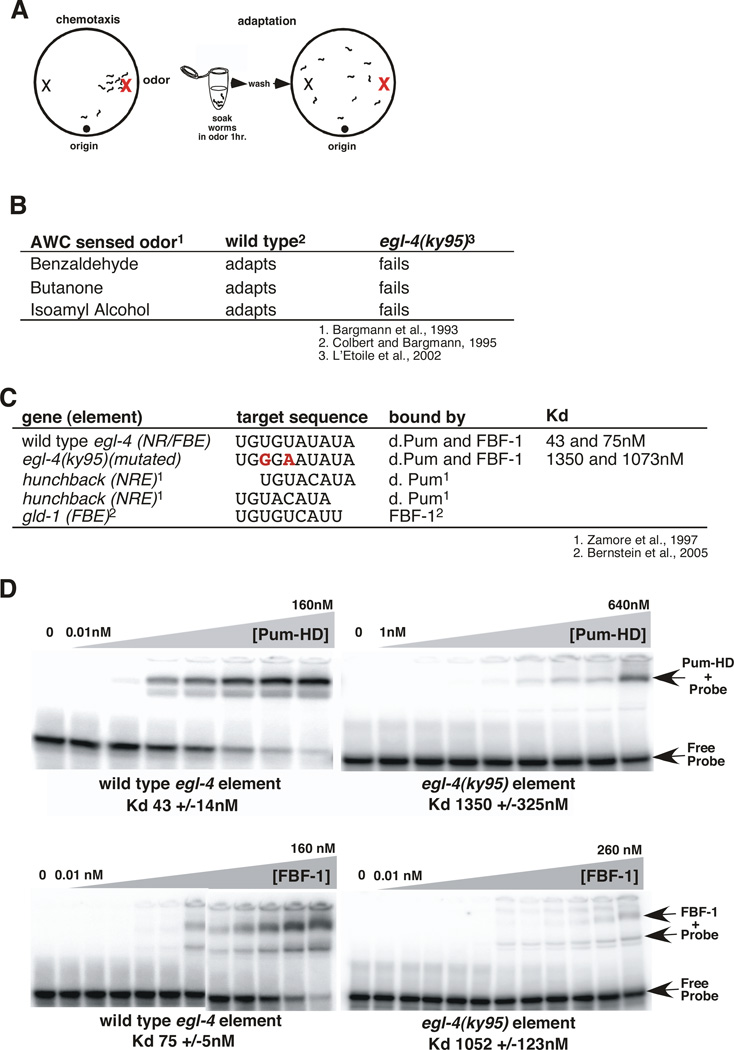
Figure 1. The egl-4 3' UTR contains a composite Pumilio and FBF binding site (NR/FBE) required for olfactory adaptation.
(A) Adaptation paradigm. In the chemotaxis assay (left), naïve worms placed on an agar lined Petri dish leave their initial location (origin) to seek an attractive odor source (red X on right side of dish). In the adaptation assay (right), worms have been exposed to the odor diluted into buffer for an hour and they no longer seek its source.
(B) Adaptation to all three AWC sensed odors was impaired by two mutations within the egl-4 3'UTR; egl-4 (ky95) (L'Etoile et al., 2002).
(C) Sequence alignment of Pumilio and FBF-1 binding sites with the egl-4 NR/FBE. Target NREs and FBEs are aligned starting with the UGU and the bases mutated in ky95 are shown in bold red. Both alignments of the egl-4 NR/FBE with the hunchback NRE are shown.
(D) Gel mobility shift assays of the putative response element within wild type (left top and bottom panels) and ky95 mutant (right, top and bottom) 3'UTRs by either purified GST-drosophila Pumilio (PUM-HD) (top panels) or purified GST-FBF-1 (bottom panels). The free probe and shifted complexes are indicated with arrows. 100 fMoles of 32P labeled 30mer RNA probes were incubated with increasing amounts of protein for two hours before being gel resolved. Concentrations of protein incubated with the wild-type element were: 0.01 to 160 nM in 2 fold steps per lane. Concentrations of protein incubated with the ky95 element were 0.01, 0.05, 0.1, 0.5, and 1 nM, then doubled beginning at 5 nM per lane; the 0.01, 0.05, and 0.1 points were omitted for the ky95 probe. The Kds below each gel were generated from three separate gel-shift assays using non-linear regression analysis (GraphPadPrism™) software to fit the % probe bound to sigmoidal dose response (variable slope) curves. Error bars indicate the s.e.m. from at least 3 separate gel shifts.