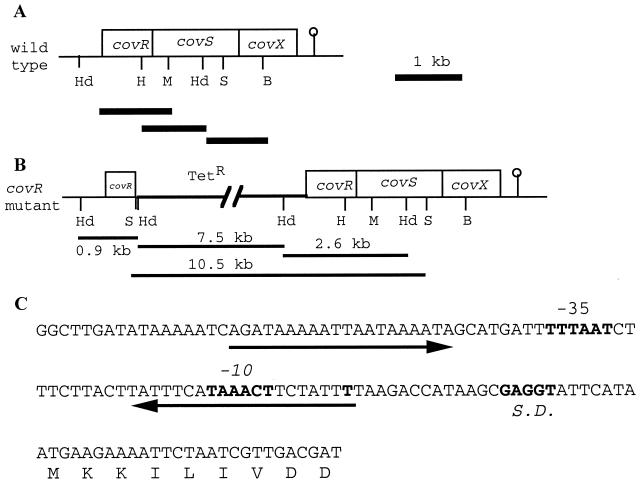
FIG. 1.
covRSX operon of S. mutans NG8. (A) Organization of the genes within the operon. The restriction sites are HindIII (Hd), HpaI (H), MscI (M), SalI (S), and BglII (B). The lollipop indicates the putative Rho-independent terminator. The thick lines under the operon are the fragments subcloned for sequencing. (B) cov operon with the insertion of pVA981 (Tetr) in covR. The 10.5-, 7.5-, 2.6-, and 0.9-kb DNA fragments observed in Southern hybridization are indicated. The 10.5- and 7.5-kb fragments were generated because of the SalI and HindIII sites from pVA981. (C) Promoter region of the operon. Arrows indicate inverted repeats. The predicted transcriptional start site (T), Shine-Dalgarno (S.D.) sequence, and −10 and −35 sequences are shown in bold type.